RNA-seq Analysis Validates the Suitability of Peptone-free McCoy’s 5A Media for Cell Culture
In this application note, we used RNA-seq and bioinformatics analysis to confirm that our new McCoy's 5A Medium, Peptone Free did not significantly affect the signaling pathways and/or genes that regulate cell growth and survival in the cell lines tested.
MoreAccelerate Cell-Based Assays with the ThawReady™ THP-1 NF-κB-Luc2 Reporter Line
In this application note, we showcase the development of a ThawReady™ THP-1 NF-κB-Luc2 cell line and demonstrate that it consistently exhibits high post-thaw viability and maintains the desired functionality.
MoreDevelopment of a ThawReady™ THP-1 Product for Cell-Based Assays
In this application note, we showcase the development and validation of an assay-ready cell product.
MoreDevelopment and Performance Evaluation of MicroQuant™
In this application note, we demonstrate the performance of MicroQuant™ in streamlining microbial quality control testing.
MoreExploring the Transcriptional Landscape of HEK Cell Lines via ATCC Cell Line Land
Our study underscores the pivotal role of RNA sequencing in unlocking the latent potential of human and mouse cell lines used in drug development, biotherapeutics production, and viral vector production.
MoreA Novel NIH/3T3 Clonal Derivative: NIH/3T3.2 Re-Developed as a Cell Model for Contact Inhibition Research
This study showcases the development of a NIH/3T3 clonal derivative that maintains the original contact inhibition characteristics associated with this cell line.
MoreDevelopment and Performance Evaluation of a Modified ABP-free McCoy's 5A Medium Formulation
Our newly developed animal by-product free McCoy’s 5A medium was tested across several ATCC cell lines for performance and was found to be a suitable substitute for the current McCoy’s 5A Medium.
More Application note
Application note
GAS-LUC2 Reporter Cell Lines for Immune Checkpoint Drug Screening in Solid Tumors
This study demonstrates the use of three robust, responsive, and reproducible GAS-Luc2 reporter solid tumor cell lines for the assessment of T cell and other immune/tumor microenvironmental cell-mediated immune responses triggered by PD-1/PD-L1, CD-155/TIGIT, or B7-H3 (CD276) immune checkpoints.
More Application note
Application note
Application of a BHK-21 gDNA Control in the Detection of Residual Host Cell DNA in Biopharmaceuticals
In this application note, we demonstrate the application of our high-quality quantitative gDNA control material in validating quantitative PCR (qPCR) assays designed to detect residual host gDNA impurities in biologics.
More Application note
Application note
Application of a HEK-293 gDNA Control in the Detection of Residual Host Cell DNA in Biopharmaceuticals
In this application note, we demonstrate the application of our high-quality quantitative gDNA control material in validating quantitative PCR (qPCR) assays designed to detect residual host gDNA impurities in biologics.
More Application note
Application note
Application of an MDCK gDNA Control in the Detection of Residual Host Cell DNA in Biopharmaceuticals
In this application note, we demonstrate the application of our high-quality quantitative gDNA control material in validating quantitative PCR (qPCR) assays designed to detect residual host gDNA impurities in biologics.
More Application note
Application note
Application of an MRC-5 gDNA Control in the Detection of Residual Host Cell DNA in Biopharmaceuticals
In this application note, we demonstrate the application of our high-quality quantitative gDNA control material in validating quantitative PCR (qPCR) assays designed to detect residual host gDNA impurities in biologics.
More Application note
Application note
Application of an Sf9 gDNA Control in the Detection of Residual Host Cell DNA in Biopharmaceuticals
In this application note, we demonstrate the application of our high-quality quantitative gDNA control material in validating quantitative PCR (qPCR) assays designed to detect residual host gDNA impurities in biologics.
More Application note
Application note
Application of a Vero gDNA Control in the Detection of Residual Host Cell DNA in Biopharmaceuticals
In this application note, we demonstrate the application of our high-quality quantitative gDNA control material in validating quantitative PCR (qPCR) assays designed to detect residual host gDNA impurities in biologics.
More Application note
Application note
3T3-L1 Cells Lose Contact Inhibition and Adipogenic Potential When Maintained in Culture at Confluence
This study determines if 3T3-L1 adipocyte differentiation is dependent on contact inhibition and tests the OP9 cell line as a possible substitute for 3T3-L1 cells in adipocyte assays.
More Application note
Application note
ATCC Minis Support VITEK 2 Quality Control Testing
This study demonstrates the use of ATCC Minis as quality control strains for microbial identification on the VITEK 2 platform.
More Application note
Application note
A Simple and Rapid Alternative to 51Chromium or Fluorescent Dye Loading for Quantification of Natural Killer Cell Activity
A flow cytometry-based assay using K-562 cells stably expressing green fluorescent protein to quantify the cytotoxic activity of primary human natural killer cells.
More Application note
Application note
ATCC Human Bronchial-Tracheal Epithelial Cells
This study will test the ability of primary human bronchial/tracheal epithelial cells (HBECs) to differentiate into mature airway tissue using an air-liquid interface (ALI) culture model.
More Application note
Application note
ATCC Strains with Demonstrated Biocatalytic Ketone Reduction Capability
This study demonstrates keto-reductase enzyme activity amongst a group of 26 ATCC strains and the potential use of this activity in industrial applications aimed at outperforming traditional chemical-based approaches.
More Application note
Application note
Comparing Toxicological Responses Between ALI-Incubated Primary Airway Epithelial Cells and Undifferentiated Counterparts
In this application note, we show that self-constructed airway models may serve as useful tools future airway toxicity research.
More Application note
Application note
Comprehensive Gene Expression Analysis and Neurotoxicity Testing of Human iPSC-derived Neural Progenitor Cells and Neurons
This study examines the ability of a novel medium to stimulate neural progenitor cells to differentiate into multiple types of neurons.
More Application note
Application note
Development and Use of Synthetic Molecular Standards for Dengue Virus Serotypes 1-4
ATCC has developed 4 quantitated synthetic molecular standards for Dengue virus (DENV) serotypes 1-4 for use as positive controls in qRT-PCR assays. Read this study to explore the development and application of these molecular standards.
More Application note
Application note
Development and Validation of a Quantitative Synthetic Molecular Standard for Monkeypox virus
In this study, ATCC designed and developed a quantitative synthetic molecular standard for Monkeypox virus to serve as a safe and reliable positive control material when conducting diagnostics assays.
More Application note
Application note
Directed Differentiation of Gastrointestinal Epithelial Organoids
Three human iPSC lines were tested for their ability to generate gastrointestinal organoids using defined culture media in a 2-D/3-D culture system using ATCC Cell Basement Membrane.
More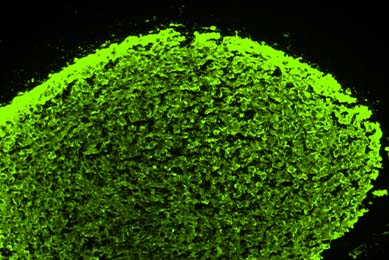 Application note
Application note
Establishment of 3-D Neurosphere Culture from Human iPSC-Derived Neural Progenitor Cells
This report describes a straightforward and robust method of generating neurospheres from neural progenitor cells
More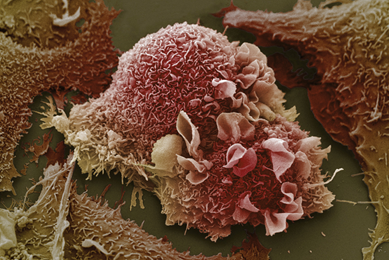 Application note
Application note
Generation of an EML4-ALK Fusion NSCLC Isogenic Cell Line Relevant for Drug Discovery and Development
An isogenic cell line was created to model cancer patients with the echinoderm microtubule-associated protein-like 4 (EML4)-anaplastic lymphoma kinase (ALK) fusion oncogene, a key oncogenic driver, and then tested for its sensitivity to known selective inhibitors of ALK.
More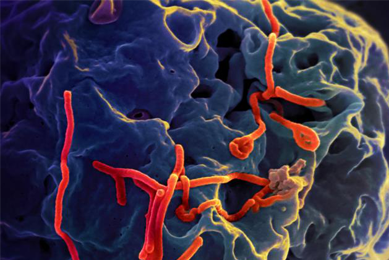 Application note
Application note
Generation of Cell Lines Capable of Producing High-titer Viral Stocks for Use in Vaccine Manufacture and Gene Therapy
In this study, we used CRISPR/Cas9 gene-editing technology to create cell lines capable of producing model clinical viruses and adeno-associated viruses at titers much higher than the parental cell lines, providing an efficient method for biopharmaceutical companies to increase production while reducing associated costs.
More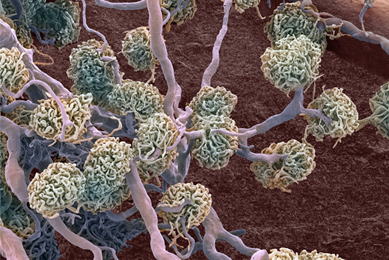 Application note
Application note
Genetically Modified Renal Proximal Tubule Epithelial Cells
We created three different hTERT-immortalized human primary renal proximal tubule epithelial cells (RPTEC) that stably express the OAT1, OCT2, and OAT3 proteins. We then tested the ability of these cells to intracellularly convey known SLC protein substrates.
More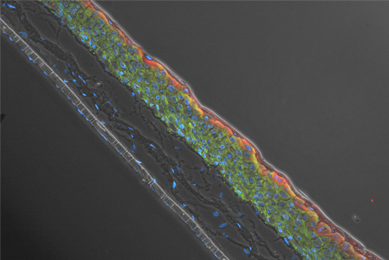 Application note
Application note
hTERT-immortalized and Primary Keratinocytes Differentiate into Epidermal Structures in 3-D Organotypic Culture
This study shows that an immortalized keratinocyte cell line, Ker-CT, is able to differentiate into organotypic skin equivalents in a 3-D culture model
More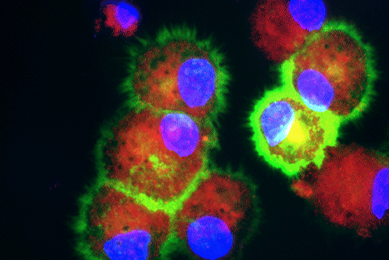 Application note
Application note
In Vitro Differentiation of Macrophages and Dendritic Cells
This study will demonstrate the differentiation potential of cryopreserved primary human CD14+ monocytes towards macrophage and dendritic cell lineages for applications including cancer immunology and vaccine development,
More Application note
Application note
Microbial Genome Databases
As part of our initiative to enhance the authentication of our products, we identified the key challenges regarding existing microbial genome databases and developed a solution for improving the quality of reference genome sequences.
MoreNovel Fluorescent Reporters for Studying Host-pathogen Interactions
We have developed green fluorescent protein (GFP)-labled strains of various Gram-negative opportunistic pathogens from the ATCC collection. Here, we demonstrate the use of these labeled strains in studying bacterial pathogenesis, host-pathogen interactions, and compound screening assays.
More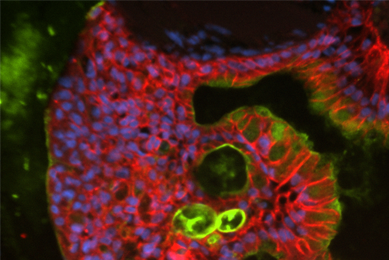 Application note
Application note
Protocol for the Thawing, Expansion, and Cryopreservation of Mouse Small Intestinal Organoids
Here, we describe a protocol for the initiation, culture, and subsequent cryopreservation of mouse small intestinal organoids starting from previously cryopreserved primary tissue–derived organoids.
More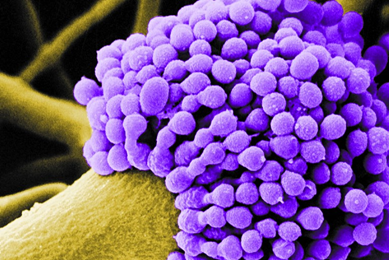 Application note
Application note
Use of the ATCC Aspergillus fumigatus Drug Testing Panel in Antifungal Drug Testing
Explore the development and application of a panel of A. fumigatus strains exhibiting various levels of sensitivity to common antifungal drugs.
More Application note
Application note
Use of Quantitative Mycoplasma DNA Controls in Evaluating Sensitivity
Learn how ATCC-designed quantitative mycoplasma DNA controls can be used to evaluate the sensitivity of two different molecular-based detection systems.
More Application note
Application note
Evaluating Airway ALI Model Fabrication Methods
This report showcases an optimal method of fabricating airway models consisting of human bronchial tracheal epithelial cells grown under air-liquid interface (ALI).
More Application note
Application note
Evaluation of GFP-labeled Control Strains for Shiga Toxin-producing Escherichia coli STEC Assays
In this study, we evaluate the use of green fluorescent protein (GFP)-labeled strains representing several clinically relevant Shiga toxin-producing Escherichia coli (STEC) serogroups associated with foodborne disease as positive controls.
More Application note
Application note
Development and Evaluation of Whole Cell- and Genomic DNA-based NGS Standards
In this study, we demonstrate that ATCC NGS Standards combined with the One Codex data analysis module provide a comprehensive solution for standardizing data from a wide range of sources, and generating consensus among microbiome applications and analyses.
More Application note
Application note
Development of ATCC Molecular Standards for Human Herpesvirus
In this study, we demonstrate the utility of molecular standards for Human herpesvirus 1 and 2 in the generation of standard curves for the detection and quantification of viral load.
More Application note
Application note
Development of Improved Synthetic Molecular Standards for Norovirus Genogroups I and II
Explore the use of quantitative synthetic molecular standards for Norovirus genogroups I and II in assay development.
More Application note
Application note
Development of Synthetic Molecular Standards for Hepatitis B and Hepatitis C Virus
Explore the development and validation of quantitated synthetic molecular standards for Hepatitis B virus (HBV) and Hepatitis C virus (HCV) and their applications as positive controls in molecular-based assays.
More Application note
Application note
Differentiation and expansion of hematopoietic precursor cells from bone-marrow derived CD34+ progenitors
This study will examine the differentiation and expansion potential of bone marrow-derived (BM) CD34+ cells into erythroid cells, megakaryocytes, and myeloid precursor cells in a liquid, non-adherent culture format.
More Application note
Application note
Development and Evaluation of Next-generation Sequencing Standards for Virome Research
In this study, we describe the development and quality control of the ATCC virome standards and present application data demonstrating their use throughout the different stages of a typical virome analysis workflow.
More Application note
Application note
Establishment and Characterization of a Kidney-drug Interaction Model
In this study, we created a HEK293T/17 cell line that stably expresses the human organic anion transporter (OAT1) and tested its transport capabilities.
More Application note
Application note
Luciferase Reporter Cancer Cell Lines Facilitate CAR-T Development
This report describes ATCC luciferase reporter solid and liquid tumor cell lines that express tumor antigens such as CD19, CD20, and HER2. Learn how you can incorporate these target cells into your T-Cell cytotoxicity assays to enable your breakthroughs in immuno-oncology.
More Application note
Application note
Telomerase-immortalized Microvascular Endothelial TIME Cells
In this study, we demonstrate that TIME cells maintain their normal growth rate and endothelial character at late passage when grown in ATCC’s VCBM-VEGF medium, but not in the previously recommended Lonza EGM™-2 MV BulletKit medium.
More Application note
Application note
Tips for Successful Subculturing: Dissociation Method
Discover how to tailor your dissociation method to your cell line to ensure successful subculture.
More Application note
Application note
Universal Mycoplasma Detection Kit
To facilitate the rapid and reliable detection of Mycoplasma in cell culture, ATCC offers the PCR-based Universal Mycoplasma Detection Kit. Explore how to use the kit to check your cultures for mycoplasma contamination.
More Application note
Application note
Use of Recombinant Bacteria with Unique Tags as Spike-in Controls for Microbiome Studies
In this application note, we describe the construction and application of ATCC Spike-in Standards as a tool for quantitative metagenomic analysis.
More Application note
Application note
Vibrio campbellii Quorum Sensing
This study will demonstrate the use of the ATCC Vibrio campbellii Panel as a non-pathogenic model for AI-2-based quorum sensing pathways.
MoreView additional resources
Culture Guides
Download these useful guides and start with fresh authenticated cells and strains from ATCC to achieve the best results.
Read the Guides
Webinars
Watch our expanding collection of webinars to learn more about the innovative research and development being performed by thought leaders in science.
Watch the WebinarsWhite Papers
Download our white papers to explore a variety of topics ranging from antimicrobial resistance to cancer research.
Read the White Papers