Authors: Helen Christina, MS, and Dev Mittar, PhD
Introduction
Development of the molecular standards
Generation of standard curves using the quantitative HSV-1 and HSV-2 molecular standards
Utility of ATCC Quantitated Molecular Standards as controls
Conclusions
References
Abstract
ATCC has developed molecular standards for Human herpesvirus 1 and 2 (HSV-1 and HSV-2) for use as controls in quantitative polymerase chain reaction (qPCR) assays. Here, we demonstrate their utility in the generation of standard curves for the detection and quantification of viral load.
Download a PDF of this application note
Download NowIntroduction
HSV-1 and HSV-2 cause a wide range of clinical manifestations that result in lifelong infections. There is currently no commercially available vaccine to protect against infection; though, antiviral medications are available for aiding in the prevention or shortening of outbreaks.1 Thus, it is imperative that infections are diagnosed early to ensure that proper treatment is provided, in turn reducing the likelihood of transmission and minimizing the frequency and severity of outbreaks.
Presently, qPCR assays are routinely used for the detection of HSV-1 and HSV-2 infections in clinical samples. However, the accuracy of a qPCR assay is dependent upon the generation of a standard curve using a positive control with a known genome copy number. Moreover, an independent positive control is required by clinical laboratories to monitor variations in assay performance for molecular assays.
ATCC offers genomic molecular standards for HSV-1 and HSV-2 for use as controls in assays designed for the detection of these viruses from clinical samples, and has recently developed a quantitative format for these standards for use in qPCR assays to detect and quantify HSV-1 and HSV-2 from unknown samples. Here, we demonstrate the utility of the quantitative molecular standards as controls in qPCR assays through the generation of standard curves for each, which were then used to quantify the working reagents for HSV-1 and HSV-2 from the National Institute for Biological Standards and Control, UK (NIBSC) as a proof-of-concept.
Development of the molecular standards
Molecular standards were prepared by extracting DNA from HSV-1 (McIntyre strain; ATCC VR-539) and HSV-2 (MS strain; ATCC VR-540) propagated in Vero cells (ATCC CCL-81). The absolute genome copy number for each molecular standard was determined using Droplet Digital PCR (ddPCR; Bio-Rad) (Table 1).
Table 1. ATCC Molecular Standards
| ATCC number | Description |
|---|---|
| VR-539DQ | Quantitative Genomic DNA from Human herpesvirus 1 (HSV-1) |
| VR-540DQ | Quantitative Genomic DNA from Human herpesvirus 2 (HSV-2) |
Generation of standard curves using the quantitative HSV-1 and HSV-2 molecular standards
Standard curves were generated using serial ten-fold dilutions of the respective quantitative molecular standard DNA, ranging from 2 copies to 2 × 105 copies/reaction for HSV-1 and for HSV-2 (Figure 1). qPCR assays were performed in triplicate using the CFX96 Real-Time PCR Detection System (Bio-Rad). The primer and probe sets from a published qPCR assay2 were used, and the cycling conditions were 50°C for 2 min, 95°C for 2 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 30 sec. The relative fluorescence unit (RFU) baseline threshold was set automatically and genome copy numbers were calculated using CFX Manager 3.0 Software (Bio-Rad).
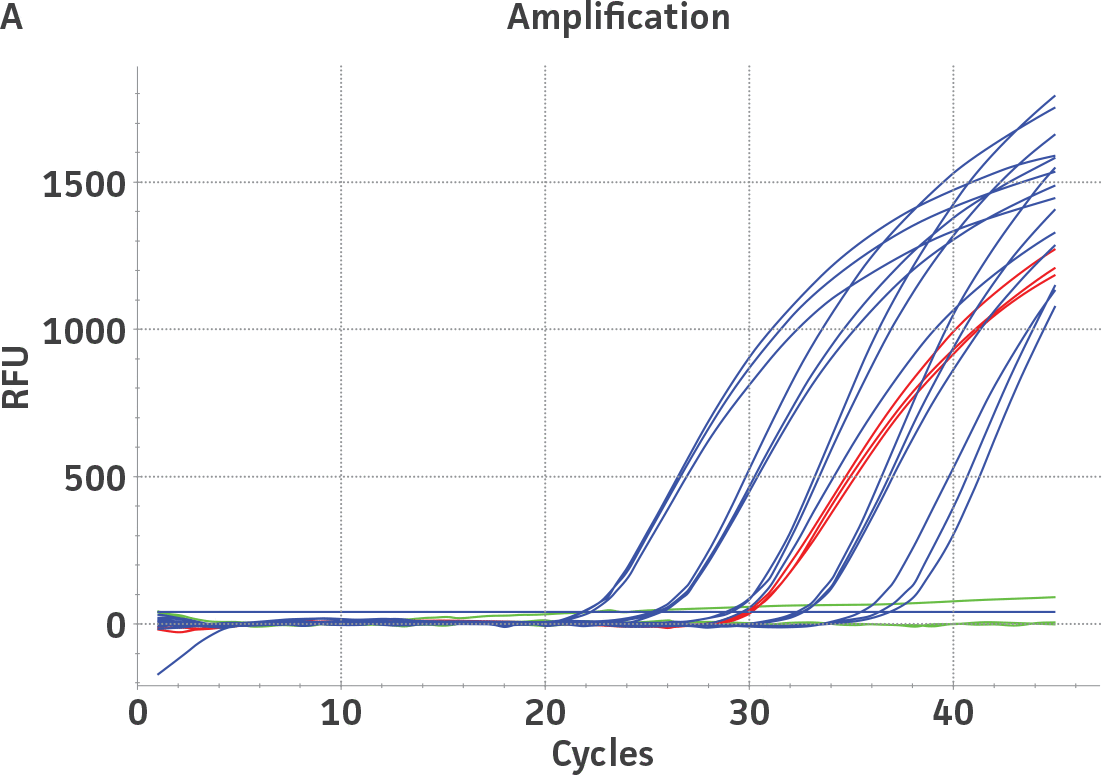
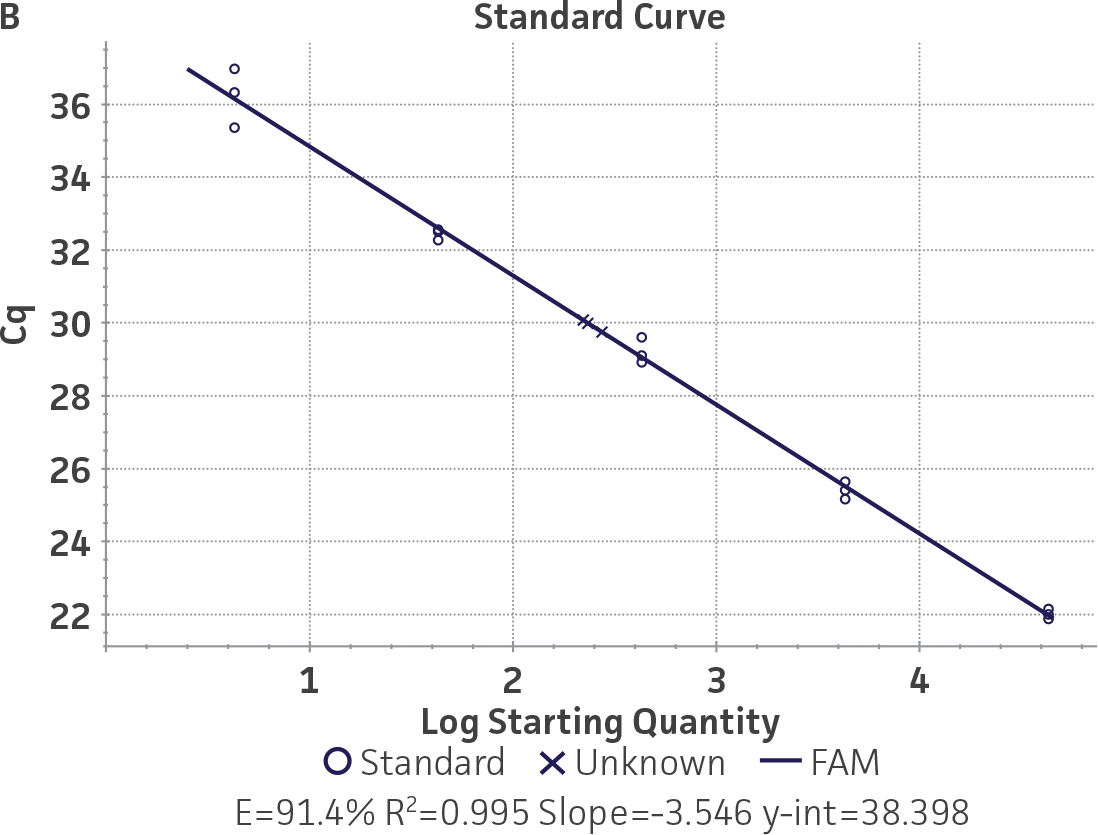
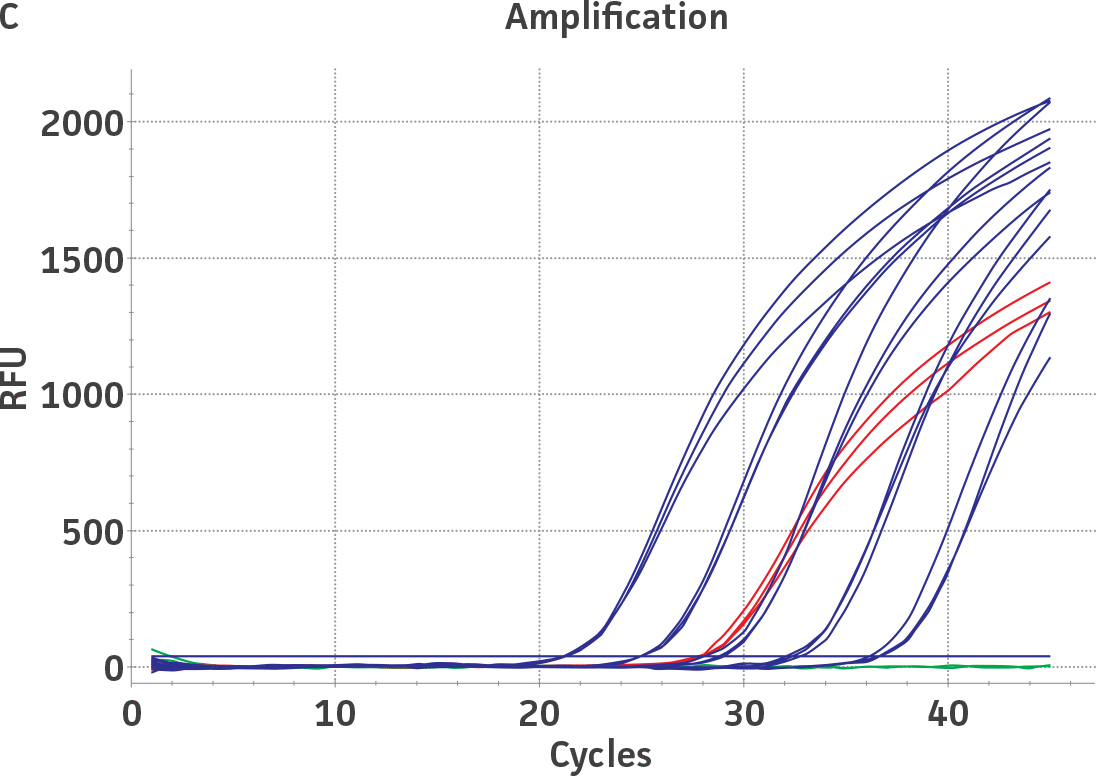
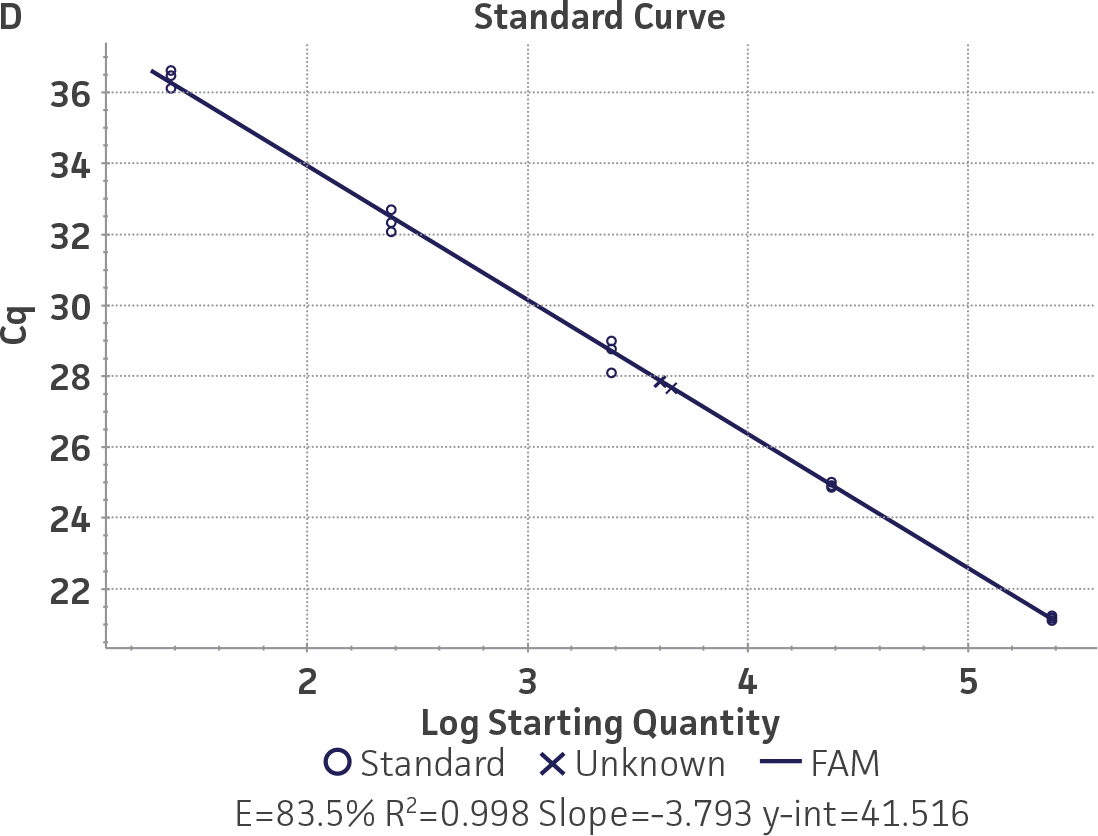
Figure 1. Generation of standard curves from ATCC quantitative molecular standards for HSV-1 and HSV-2. (A and C) Amplification plots and (B and D) standard curves generated using the HSV-1 and HSV-2 quantitative molecular standards (ATCC VR-539DQ and ATCC VR-540DQ, respectively) with the primer and probe set from the published qPCR Assay.2 (Blue) = Serial ten-fold dilutions of the quantitative molecular standard. (Green) = Negative control.
Utility of ATCC Quantitated Molecular Standards as controls
The working reagent for HSV-1 and HSV-2 (NIBSC, UK) were quantified using the standard curves generated from the quantitated HSV-1 and HSV-2 molecular standards, respectively (Figure 2). Viral DNA from the HSV-1 and HSV-2 working reagents was extracted using the QIAamp Viral RNA Mini Kit (QIAGEN) and subsequently diluted for quantification by qPCR. Both working reagents were run in triplicate wells (n=3) using the HSV-specific published assay.2 The average from triplicate wells were used to calculate the quantities of the HSV-1 and HSV-2 genomes, respectively. Error bars indicate standard deviation and were calculated using GraphPad Prism software.
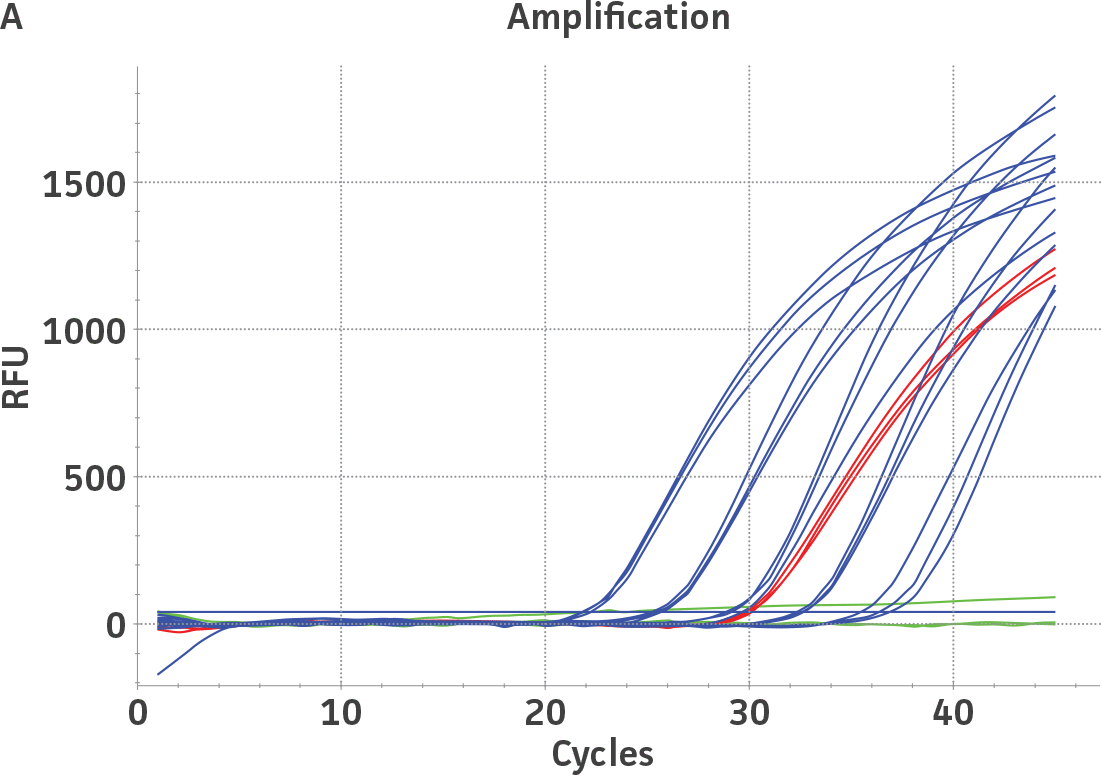
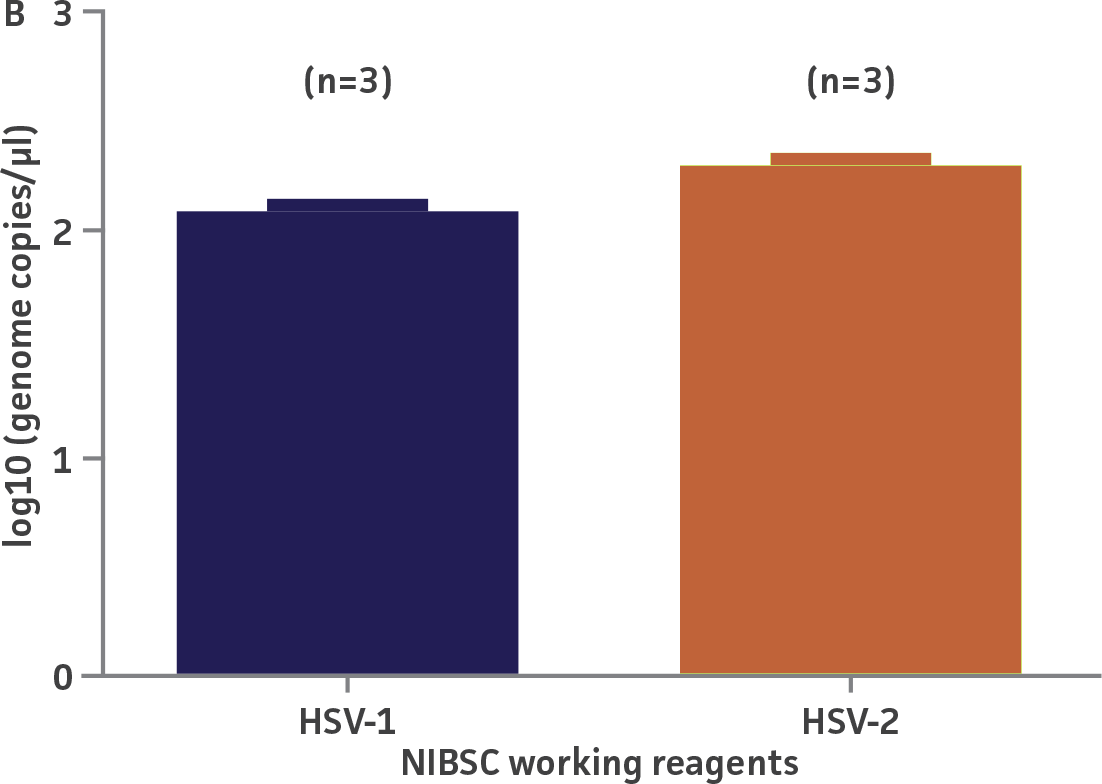
Figure 2. Quantification of NIBSC working reagents for HSV-1 and HSV-2 using ATCC quantitative molecular standards. (A) An example of a qPCR amplification plot showing the ATCC quantitative molecular standards (Blue), the NIBSC working reagent for HSV-1 (Red), and a negative control (Green). (B) Quantitation of the working reagent for HSV-1 (Blue) and HSV-2 (Red).
Conclusions
ATCC HSV-1 and HSV-2 quantitative molecular standards provide well-characterized reference materials for qPCR assays. They are ideal for use in the detection of HSV-1 and HSV-2, and the quantitative format enables the generation of a standard curve for determining viral load. Further, these standards are compatible with published assays for HSV-1 and HSV-2, and can be used as controls for assay development, verification, and validation. Taken together, these molecular standards provide convenient, well-characterized HSV reference materials for use in molecular-based assays.
Download a PDF of this application note
Download NowReferences
- Genital Herpes - CDC Fact Sheet. (2015, November 17). Retrieved October 27, 2016, from https://stacks.cdc.gov/view/cdc/34405/
- Ryncarz AJ, et al. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J Clin Microbiol 37(6): 1941-1947, 1999. PubMed: 10325351