Authors: Shamaila Ashraf, MSc, MPhil, PhD; Afshin Sohrabi, PhD; Kurt Langenbach, PhD; and Dev Mittar, PhD
Introduction
Development of the synthetic molecular standards
Quantification of native DENV 1-4 RNA using the standard curves
Conclusions
References
Abstract
ATCC has developed four quantitated synthetic molecular standards for Dengue virus (DENV) serotypes 1-4 for use as positive controls in quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays. These synthetic molecular standards comprise short conserved fragments representing the DENV genome, and are compatible with numerous published qRT-PCR assays to determine viral load. Moreover, they can be handled at biosafety level one (BSL-1), are stable, and can be shipped internationally.
Download a PDF of this application note
Download NowIntroduction
Dengue is an acute febrile illness caused by any one of four serotypes (1-4) of genetically-related DENV.1,2 Due to its reemergence as a global disease, dengue is considered to be the most common and clinically important arbovirus disease, with an estimated 400 million cases reported annually.1,3 So far, no vaccine or antiviral drugs are available for the treatment of dengue and the only preventive measure is to control the spread of mosquitos. Therefore, the detection and surveillance of dengue is very important. Currently, qRT-PCR is the preferred method for the detection and quantification of DENV in clinical diagnostics and epidemiological surveillance. The accuracy of a qRT-PCR assay relies on the generation of a standard curve using a positive control based on known viral genome concentration. Moreover, an independent standard is required by diagnostic laboratories for verification and validation of commercial and lab-developed assays.
Native DENV RNA can be used as a standard for these assays; however, the full-length RNA is on the Commerce Control List and requires a permit from the US Department of Commerce for international shipment. To address this, ATCC has developed four quantitated synthetic molecular standards that represent DENV serotypes 1-4 (Table 1). As compared to native RNA, these synthetic standards are easier to use as controls for qRT-PCR assays, exhibit less variability, have a longer shelf life, and eliminate the need to culture viruses. These standards could be used to monitor assay-to-assay and lot-to-lot variability or as an independent standard for validation and verification studies. In the following proof-of-concept study, we demonstrate the use of the ATCC synthetic DENV molecular standards in the development of a standard curve for the quantification of viral RNA extracted from representative samples of all four DENV serotypes.
Development of the synthetic molecular standards
The DENV 1-4 molecular standards were developed using a proprietary artificial RNA synthesis method. Each standard was designed to contain short fragments from the capsid, membrane, and envelope genes of the DENV genome, as well as the target regions encompassing the primer sequences from numerous published RT-PCR assays, including the DENV 1-4 Real-Time RT-PCR Assay developed by the Centers for Disease Control and Prevention (CDC).4 The synthetic RNA standards were quantified by Droplet Digital PCR (ddPCR, Bio-Rad) in order to package precise copies of RNA. Given the inherent labile nature of RNA, a proprietary stabilization matrix was added to the quantitated RNA preparations.
Generation of standard curves from DENV synthetic molecular standards
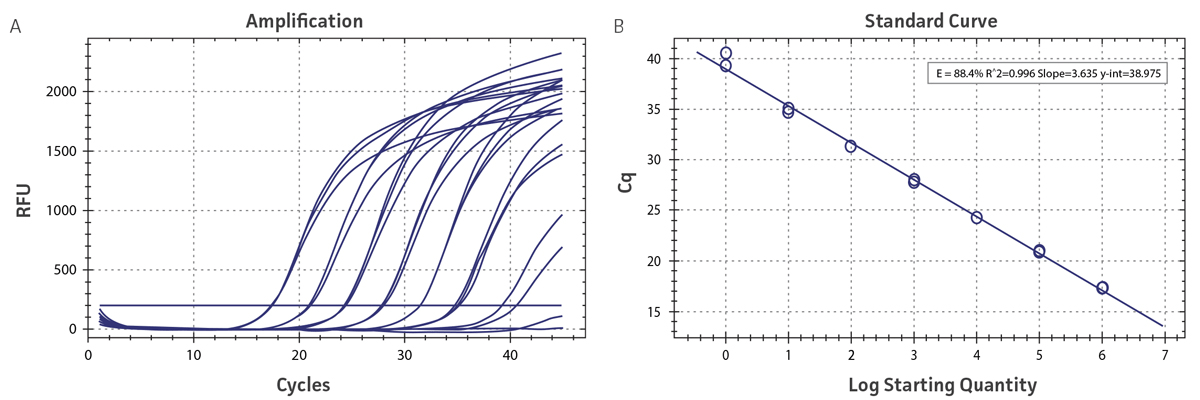
Standard curves were generated using serial ten-fold dilutions of the synthetic RNA standards, ranging from 1 copy to 1 x 106 copies/µL (Figure 1; DENV 1-3 data not shown). Standards were tested in triplicate wells using qRT-PCR according to instructions provided in the CDC Real-Time RT-PCR assay,4 with slight modifications, using the CFX96 Real-Time PCR Detection System (Bio-Rad). The following primer sets were employed to examine each individual dengue serotype for compatibility: the primer set included in the CDC Real-Time RT-PCR assay4 and the primer set published by Waggoner et al.5 Cycling conditions for all primer sets were 50°C for 15 min and 95°C for 2 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 30 sec. The exception was for the DENV-4 Waggoner et al. primer set, where the annealing temperature was 55°C for 30 sec. The relative fluorescence unit (RFU) baseline threshold was calculated using CFX Manager 3.0 Software (Bio-Rad). From this analysis, it was determined that each of the DENV 1-4 synthetic molecular standards is compatible with both primer sets over a broad linear dynamic range, and exhibits minimal variability as evident from slope and R2 values (Table 2).
Figure 1. Generation of standard curves using the DENV-4 Molecular Standard. (A) qRT-PCR amplification plot and (B) standard curve generated using the DENV-4 molecular standard in conjunction with the CDC primer and probe set.4
Table 2. Slope and R2 values generated using DENV 1-4 molecular standards
| Primer and probe | DENV-1 | DENV-2 | DENV-3 | DENV-4 | |
|---|---|---|---|---|---|
| CDC Assay4 | Slope | -3.244 | -3.277 | -3.315 | -3.642 |
| CDC Assay4 | R2 | 0.99 | 0.996 | 0.987 | 0.996 |
| Published Assay5 | Slope | -3.536 | -3.535 | -3.705 | -3.775 |
| Published Assay5 | R2 | 0.991 | 0.997 | 0.989 | 0.996 |
Quantification of native DENV 1-4 RNA using the standard curves
The following DENV strains from ATCC and BEI Resources, representing DENV serotypes 1-4, were quantified using the standard curves generated from the DENV 1-4 synthetic molecular standards, respectively:
DENV 1 = TH-S-man (ATCC VR-1586)
DENV 2 = New Guinea C (NR-84, BEI Resources)
DENV 3 = Philippines/H87/1956 (NR-80, BEI Resources)
DENV 4 = H241 (ATCC VR-1257)
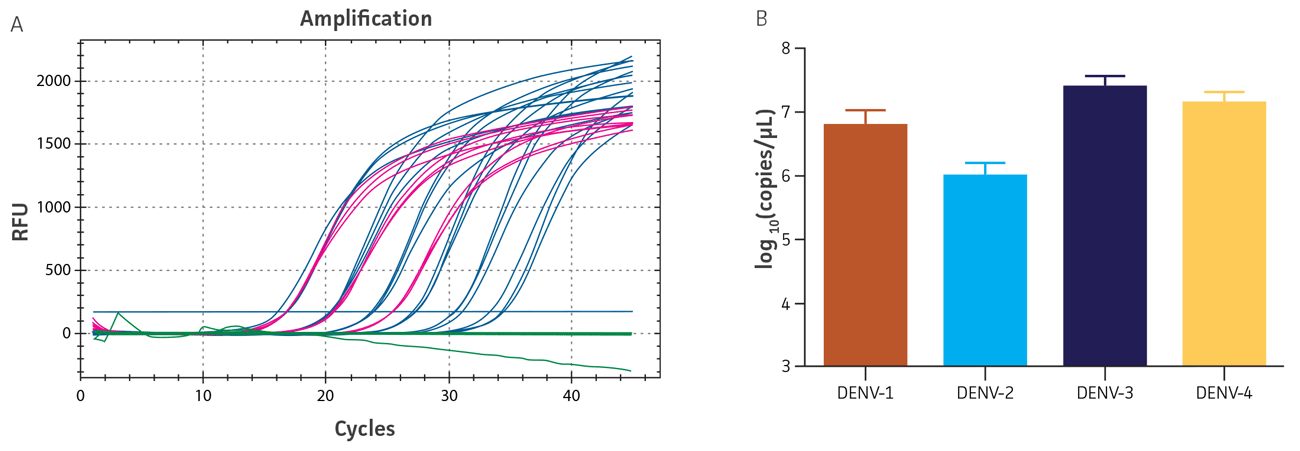
Viral RNA was extracted using the QIAamp Viral RNA Mini Kit (QIAGEN), and subsequently diluted 1:100, 1:1000, and 1:10,000 for quantification via qRT-PCR. Uninfected Vero (ATCC CCL-81) and LLC-MK2 Derivative (ATCC CCL-7.1) cell lines were used as negative controls. The titers of the native DENV 1-4 samples were determined using the respective standard curves generated by the synthetic molecular standards; the primers employed were those designed for the DENV 1-4 Real-Time RT-PCR Assay developed by the CDC (Figure 2A, DENV 1-3 data not shown; Figure 2B).
Figure 2. Quantification of native DENV RNA. (A) qRT-PCR amplification plot showing the DENV-4 synthetic molecular standard (blue), the native dengue virus sample (pink), and the negative control sample (green). (B) Titers of DENV-1, -2, -3, and -4 samples as determined by the qRT-PCR standard curves generated using the DENV 1-4 molecular standards, respectively.
Conclusions
We have generated fully authenticated and characterized synthetic molecular standards for DENV 1-4 that can be used as positive controls in qRT-PCR assays. These standards are quantitated, stable, can be handled in BSL-1 conditions, and don’t require special permits for international shipping. The DENV 1-4 molecular standards are compatible with primers from the CDC DENV-1-4 Real-Time RT-PCR Assay4 and another multiplex real-time RT-PCR assay5 over a broad linear dynamic range, and exhibited minimal variability as evident from slope and R2 values. Moreover, these standards can used to determine the viral load of unknown DENV samples from each of the four serotypes through the generation of a standard curve. Taken together, this synthetic molecular standard approach provides a convenient and well characterized DENV reference material for use in molecular-based assays, and can be further extended to other microorganisms which are unculturable or need to be grown in a high-containment facility.
Download a PDF of this application note
Download NowReferences
1. CDC. Dengue, http://www.cdc.gov/Dengue, 2013.2. Back AT, Lundkvist A. Dengue viruses - an overview. Infection ecology & epidemiology 3: doi:10.3402/iee.v3i0.19839, 2013.
3. Guzman MG, et al. Dengue: a continuing global threat. Nature reviews. Microbiology 8: S7-16, doi:10.1038/nrmicro2460, 2010.
4. CDC. DENV-1-4 Real-Time RT-PCR Assay for Detection and Serotype Identification of Dengue virus. Instructions for Use Package Insert, http://www.cdc.gov/dengue/resources/rt_pcr/CDCPackageInsert.pdf, 2013.
5. Waggoner JJ, et al. Single-reaction, multiplex, real-time RT-PCR for the detection, quantitation, and serotyping of dengue viruses. PLoS
neglected tropical diseases 7: e2116, doi:10.1371/journal.pntd.0002116, 2013.