Authors: Gaganpreet Kaur Monga, MS; Jung-Woo Sohn, PhD; Quan Zhang, PhD; Robert McLaughlin, PhD; and Neal Connors, PhD
Introduction
Results and Discussion
Methods and Materials
References
Abstract
This study demonstrates keto-reductase enzyme activity amongst a group of 26 ATCC strains and the potential use of this activity in industrial applications aimed at outperforming traditional chemical-based approaches. This activity was supported by bioinformatic analysis of whole-genome sequence data, which revealed a large number of keto-reductase enzymes. New keto-reductases can expand the range of ketone substrates that can be reduced (Figure 1) and increase the number of enzymes available for heterologous expression and protein engineering. Expanding the role of microbial enzymes in biotechnological applications, particularly in the field of asymmetric synthesis, allows for the development and realization of highly selective and efficient biocatalytic reactions on an industrial scale.
Download a PDF of this application note
Download NowIntroduction
Biocatalysis has emerged as an integral technology for the design and development of efficient processes for the pharmaceutical, bulk, and fine chemical industries. Enzyme-based catalysis meets the increasing demands for highly selective, safe, and sustainable industrial processes. Recent advances in metagenomics, genetic engineering, and bioinformatics have allowed for rapid identification and development of new enzymes that enable the transformation of pro-chiral substrates (ie, ketones) to attain yields and selectivity yet unreached. Offering both economic and environmental advantages over chemical methods, biocatalysis applies inexpensive, renewable resources, many of which are biodegradable while fulfilling key attributes of green and sustainable development.1
One of the most exploited enantioselective biotransformations in industry is the reduction of a ketone to an optically pure secondary alcohol that can serve as a building block for many pharmaceutically active compounds and specialty chemicals.2 For example, biocatalytic ketone reductions at the kg-scale have been reported for the synthesis of Brilinta (ticagrelor; a blood thinner), Olysio (simeprevir; a hepatitis C drug), and Imbruvica (ibrutinib; a lymphoma/leukemia treatment[ii]), which are pharmaceutical products with billions of dollars in annual sales. Historically, biocatalytic ketone reductions have made use of baker’s yeast (Saccharomyces cerevisiae),4 but more recently an increasing number of alternate organisms with keto-reductase activity have been identified and have been explored. The identification of additional yeast strains and other microbes capable of ketone reductions expands the scope of substrates that can be reduced beyond the capabilities of S. cerevisiae with a concomitant increase in the number and diversity of keto-reductase enzymes available.3,5 The identification of additional microbes with keto-reductase activity will further expand the range of ketone substrates that can be reduced and provide for new possibilities in protein engineering.
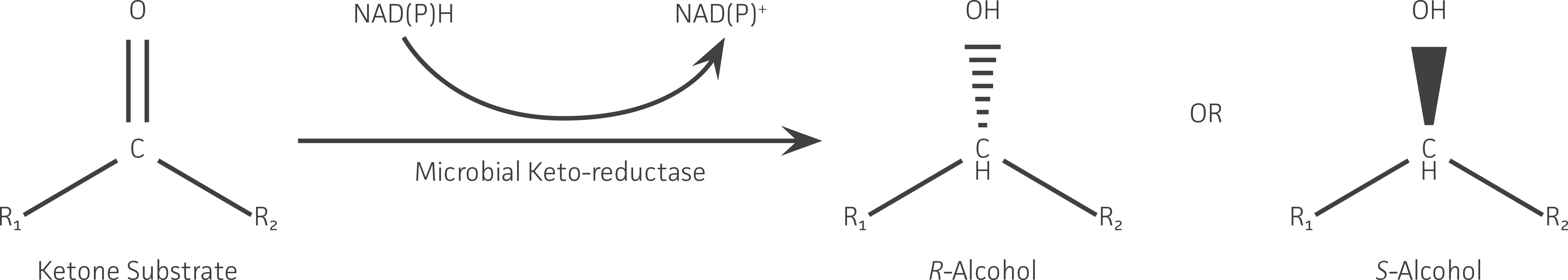
Figure 1. Representative keto-reductase reaction used in the asymmetric (ie, chiral) synthesis of a secondary alcohol from a ketone. The substrate specificity (ie, R groups that can be accepted) and the R or S stereochemistry of the alcohol product formed is determined by the enzyme’s structure.
In this study carried out at the ATCC Center for Translational Microbiology (ATCC-CTM) at the Institute for Life Science Entrepreneurship (ILSE), a collection of ATCC microbes comprising 24 fungi (20 yeast strains and 4 filamentous fungi) and 2 bacteria including 1 actinomycete (Table 1) were identified from the PubMed Central database and evaluated for keto-reductase activity against a panel of representative ketone substrates (Table 2). The keto-reductase activity demonstrated in this collection reveals the untapped potential available to parties interested in producing valuable, optically active alcohols from pro-chiral ketone substrates to serve as building blocks for pharmaceutically active compounds and specialty chemicals.
Table 1. Twenty-six ATCC strains identified from the PubMed Central database search for strains with ketone reduction capability. The list of strains includes 20 yeasts (Y), 4 fungi (F), 1 bacterium (B), and 1 actinomycete (A).
|
|
(a) Type strain of Candida guilliermondii; (b) Deposited to ATCC as Candida famata; (c) Deposited to ATCC as Penicillium marneffei.
Table 2. Representative ketone substrates and corresponding R and S or D and L alcohol products. All ketone substrates are liquids with a specific gravity of ≈1 g/mL.
| Substrate class | Substrate | Product (R-isomer) | Product (S-isomer) |
|---|---|---|---|
| Aryl ketone | 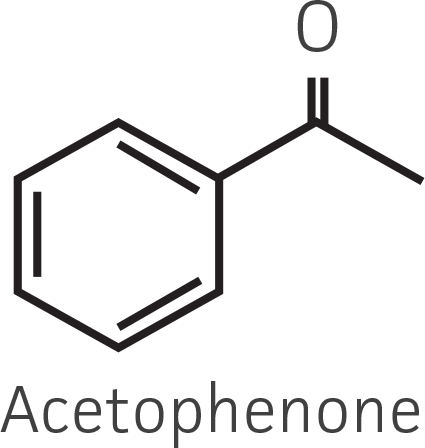 |
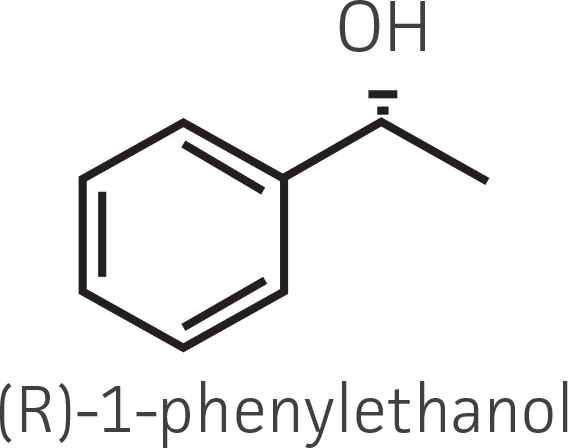 |
 |
| α-ketoester |  |
 |
 |
| β-ketoester | 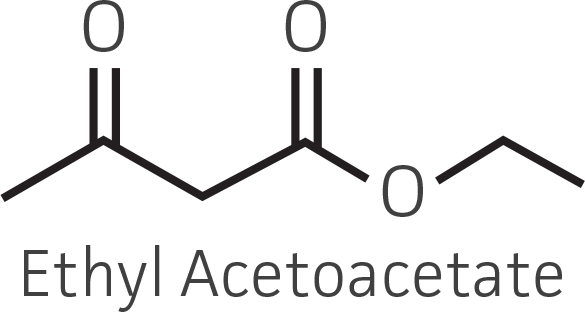 |
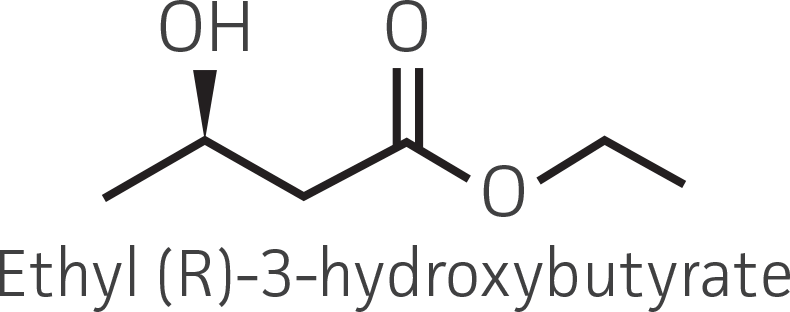 |
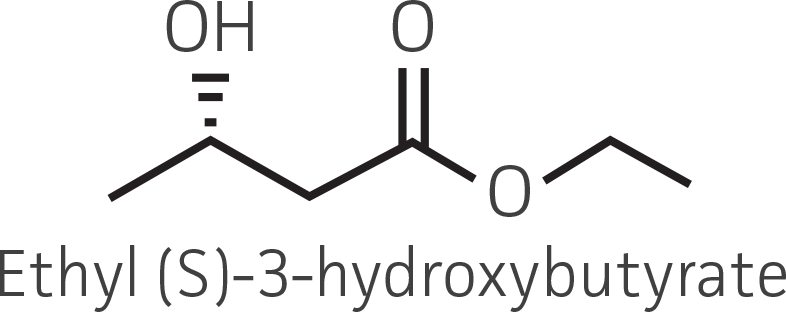 |
| β-ketoester (electron withdrawing group) | 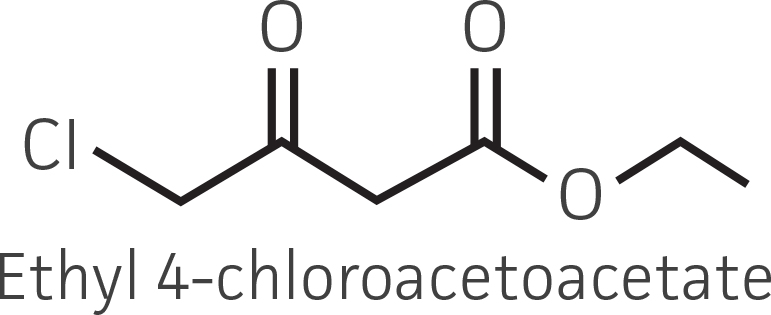 |
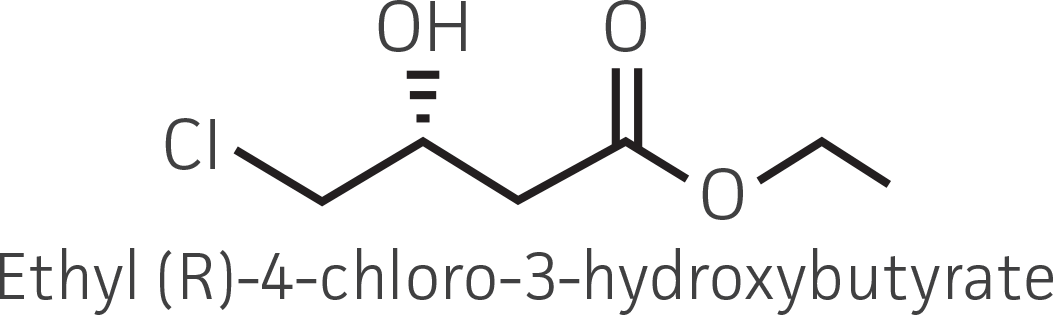 |
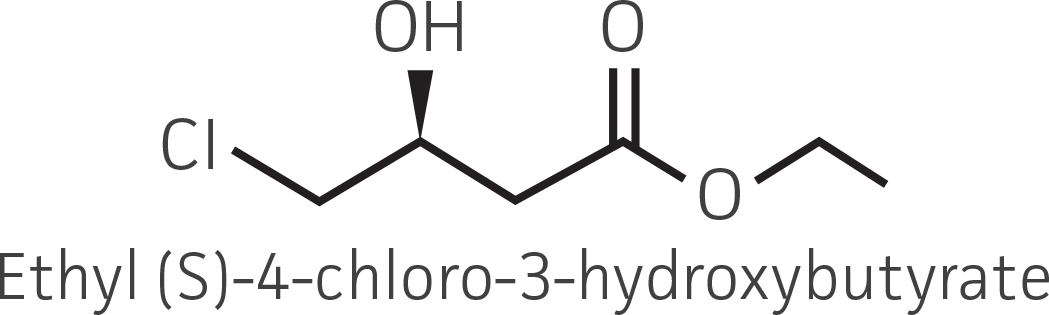 |
Results and discussion
Conversion by freshly cultured cells
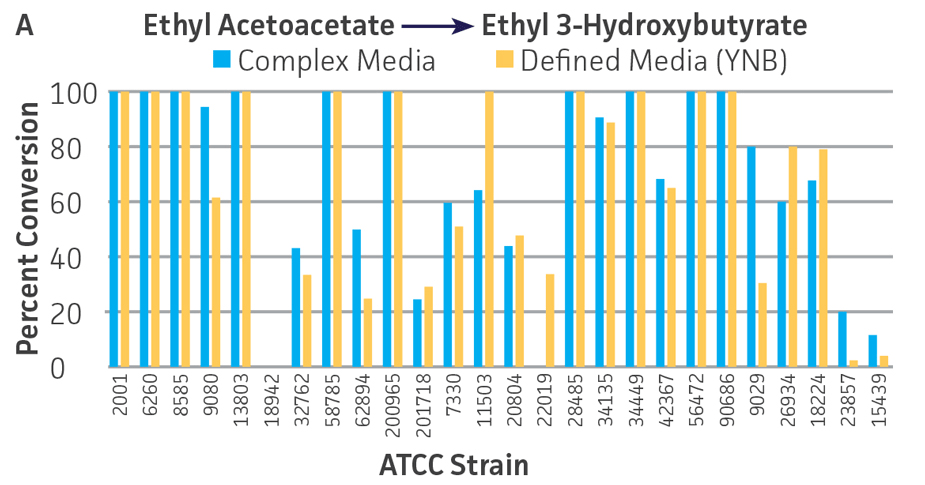
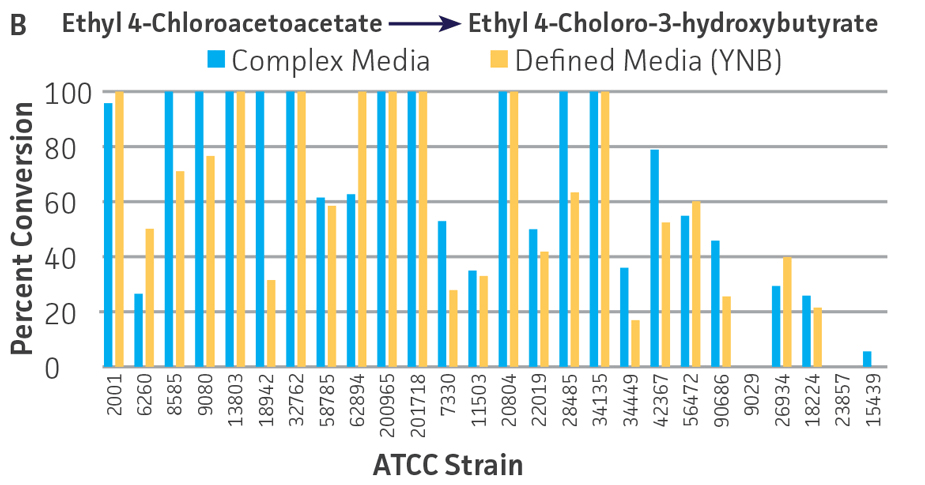
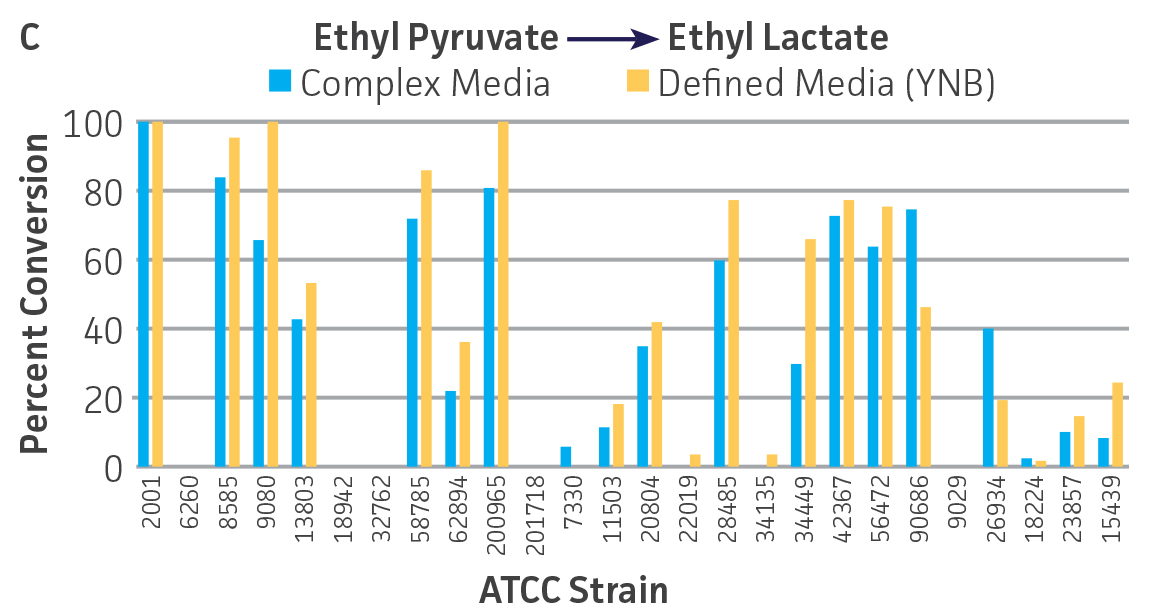
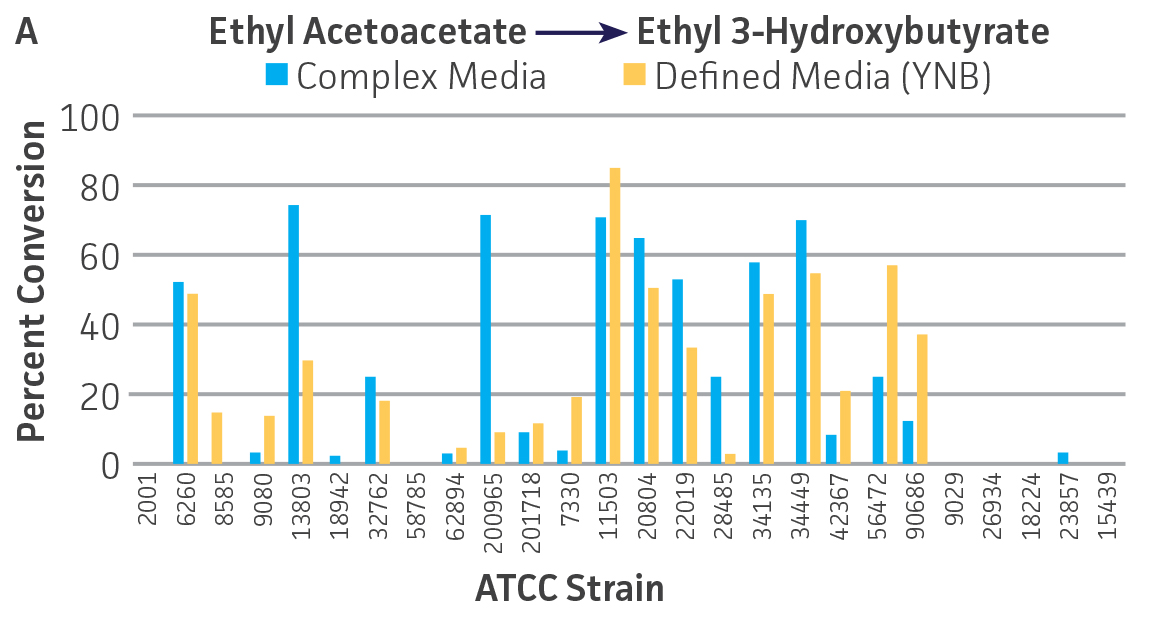
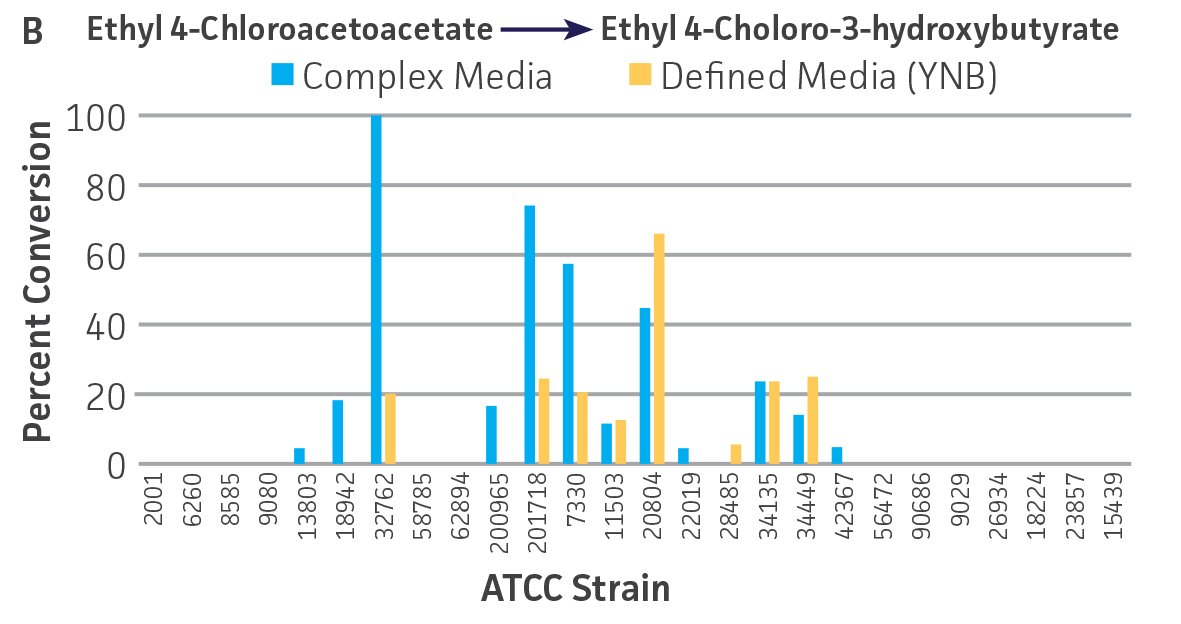
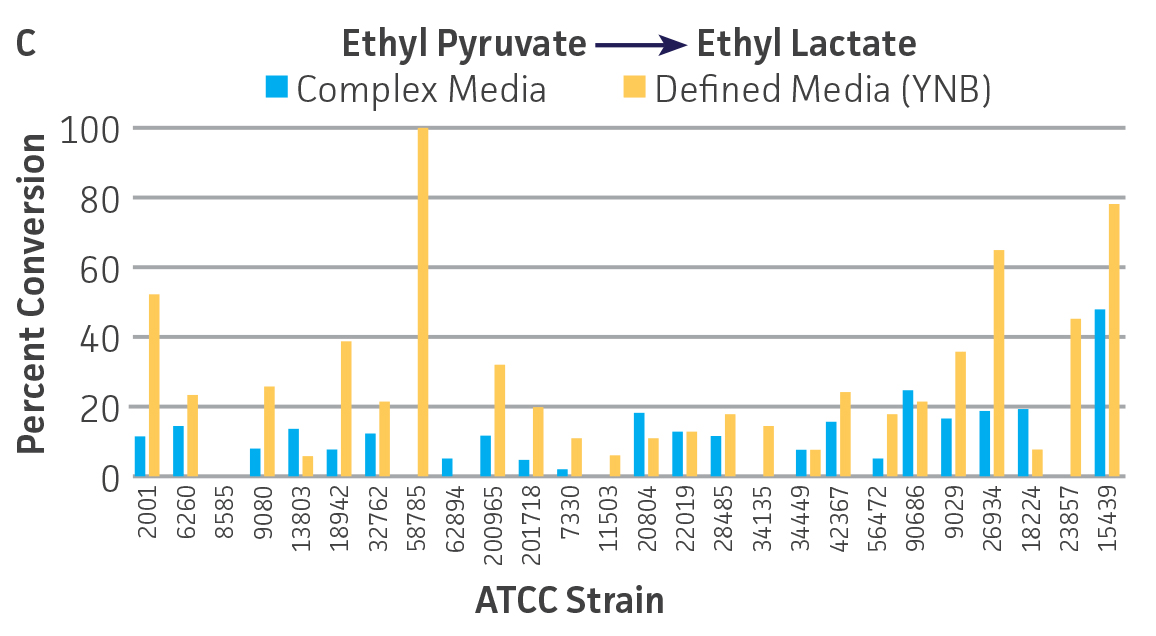
Collectively, the “freshly” grown microbes converted the various ketone substrates to different degrees, with some differences observed between complex and defined media (Figures 2 A-D). Conversion of a ketone substrate is a function of the selectivity of one or more enzymes within the cell but also the relative expression of those enzymes that can be impacted by growth media composition. In most cases, the biocatalytic activity in complex media was greater or equivalent to that observed in defined media with the noted exception of C. parapsilosis (ATCC 22019), which was able to convert ethyl acetoacetate only under defined media conditions. However, conversion based on growth in complex media increased the number of background peaks on the chromatograms, perhaps making the defined media more desirable for interpreting data for other substrates (data not shown). Other growth parameters such as temperature, pH, or primary carbon source were not explored as part of this study, although these variables can still have an impact on the extent and rate of conversion.
Interestingly, there were differences across the entire group of strains in the conversion of ethyl acetoacetate (Figure 2A) vs. the chlorinated analogue ethyl 4-chloroacetoacetate (Figure 2B). This suggests that the keto-reductases within some of the organisms can tolerate or even prefer an electron withdrawing group. None of the strains were particularly prolific at converting acetophenone to phenylethanol, though specificity of the enzymes and expression level cannot be differentiated. No attempt was made to measure enantiomeric excess (%ee) as it is generally accepted that microbes, in particular yeasts, can have more than one enzyme that can reduce ketones and the resulting %ee is a composite of these different enzymes with different stereospecificities.
 |
 |
 |
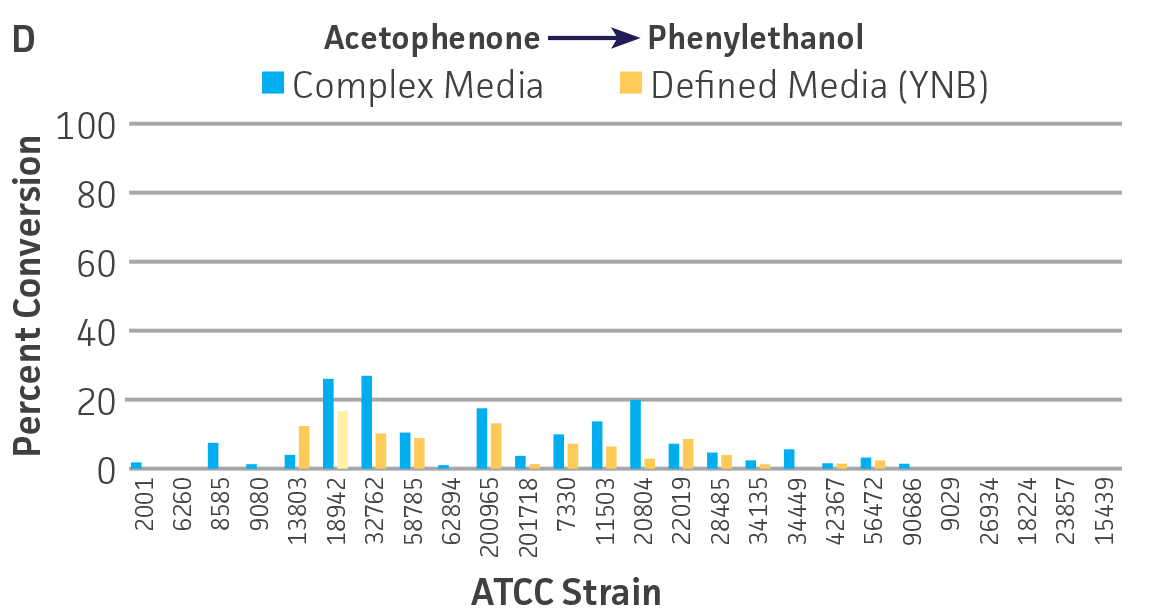 |
Figure 2. Conversion of (A) ethyl acetoacetate, (B) ethyl 4-chloroacetoacetate, (C) ethyl pyruvate, and (D) acetophenone to their corresponding alcohols by ATCC strains freshly grown and resuspended in complex or defined media.
Conversion by lyophilized cells used directly
The use of lyophilized cells allows the growth and preparation of the microbial catalyst to be temporally separated from the actual screening. Moreover, microbial catalysts can be lyophilized in a well-plate format and subsequently screened in an automated fashion along with traditional chemical catalysts. Encouraging results were obtained for the use of lyophilized microbial catalysts stored at 4°C, without a period of outgrowth after resuspension and prior to substrate addition, and suggest that a lyophilized keto-reductase “panel” can be created in deep-well plates to facilitate rapid screening without the need to carry out culturing at the time of bioconversion. Conversion capabilities for the lyophilized strains were generally reduced, and in some cases eliminated, compared to that of actively metabolizing strains (Figures 3A-D). However, there were some instances where lyophilization and direct use improved substrate conversion – eg, C. parapsilosis (ATCC 22019) / ethyl acetoacetate and S. venezuelae (ATCC 15439) / ethyl pyruvate).
The use of 10% (w/v) trehalose is a generally accepted lyoprotectant, but others along with different growth conditions and the physiological state of the cells at the time of lyophilization are known to impact viability17 and by extension, the ability to convert ketone substrates. Moreover, the impact of different preservation variables varies by strain. This is particularly useful for identifying a strain possessing a keto-reductase enzyme active against a substrate of interest that ultimately will be expressed in a heterologous host, and where the absolute conversion capability of the native strain is not as important.
 |
 |
 |
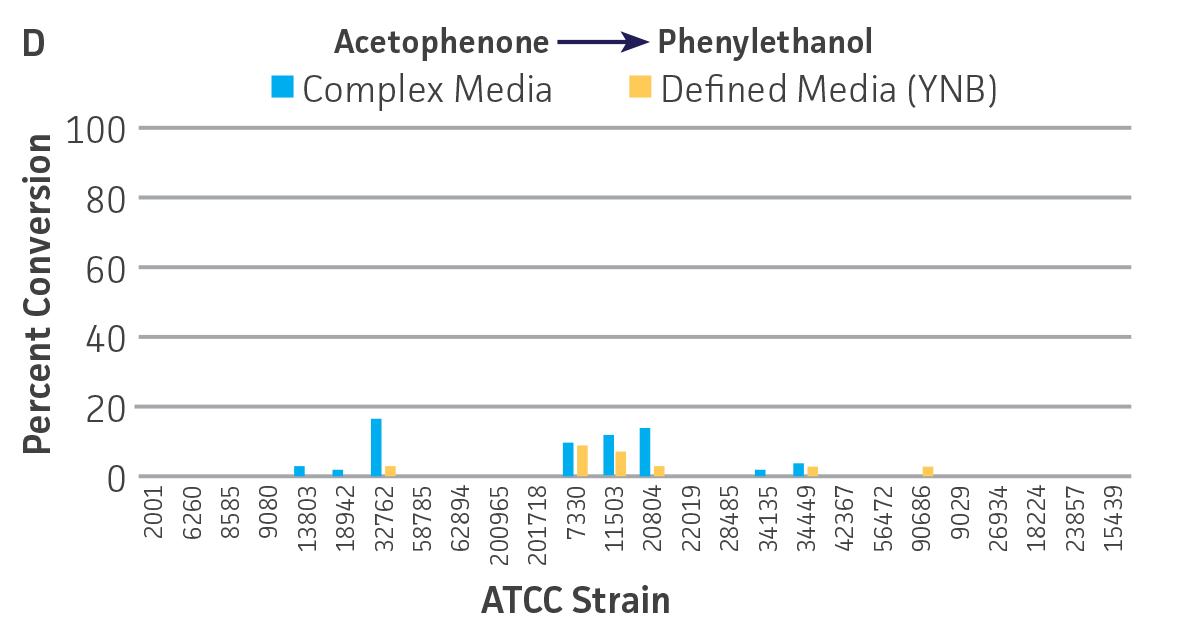 |
Figure 3. Conversion of (A) ethyl acetoacetate, (B) ethyl 4-chloroacetoacetate, (C) ethyl pyruvate and (D) acetophenone to their corresponding alcohols by ATCC strains lyophilized in complex or defined media plus 10% (w/v) trehalose, stored at 4°C.
Keto-reductase diversity of sequenced strains
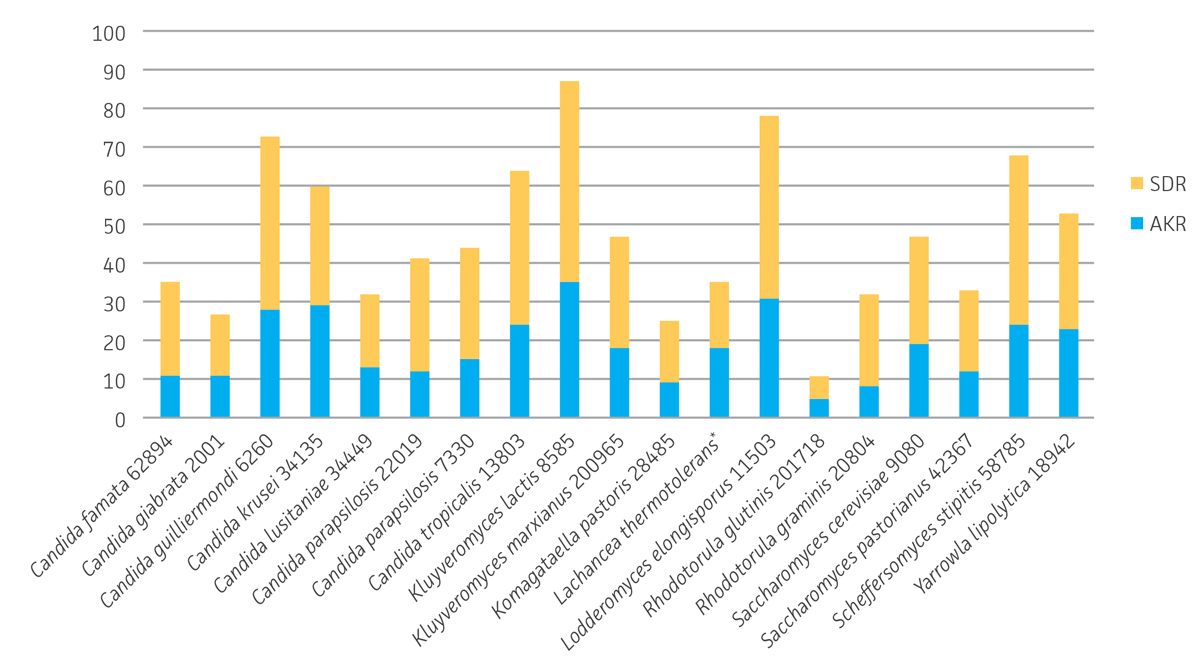
S. cerevisiae possesses several enzymes from different superfamilies capable of carrying out the reduction of ketone substrates.4 Analysis whole-genome sequence data from 19 of the yeast strains were analyzed for keto-reductase diversity based on aldo-keto reductase (AKR) and short chain dehydrogenase/reductase (SDR) enzyme superfamilies using Prosite18 and Pfam19 motifs. Based on the presence of at least one Prosite or Pfam signature, these 19 strains possess a total of approximately 345 AKRs and 547 SDRs (892 total) that are potentially capable of converting ketone substrates to alcohols (Figure 4). K. lactis (ATCC 8585) had the most AKRs and SDRs of the group with a total of 87. The Candida genus has long been a source of keto-reductase enzymes and in this study; sequencing revealed 73 total AKRs and SDRs for C. guilliermondii (ATCC 6260) while C. krusei (ATCC 34135) and C. tropicalis (ATCC 13803) possess a total of 60 and 64 respectively. Surprisingly L. elongisporus (ATCC 11503) possessed 78 total enzymes, and there has been one report of an AKR from this strain converting ethyl 4-chloroacetoacetate to the corresponding hydroxybutanoate having been cloned and over-expressed in E. coli.20 S. stipitis (ATCC 58785), known for its ability to ferment xylose to ethanol, also represents a rich source of AKR and SDR enzymes with a total of 68 enzymes. By extrapolation, these results suggest that the entire panel of strains may possess over one thousand potential keto-reductases. While duplication of enzymes is very likely, this simple analysis implies an extensive amount of keto-reductase enzyme diversity to be explored for conversion of novel substrates.
ATCC Strains
Figure 4. Identification of aldo-keto reductase (AKR) and short chain dehydrogenase/reductase (SDR) enzyme superfamily members in whole-genome sequenced yeast strains. *Based on GenBank Reference GCA_000142805.1
Methods and materials
An ILSE-developed algorithm was used to search the PubMed Central (PMC) literature database, revealing 40 ATCC cultures that have previously been used or screened for catalyzing the reduction of ketones to secondary alcohols. A shortlist of 20 yeasts, 4 fungi, 1 actinomycete, and 1 bacterium (Table 1) was ultimately selected and evaluated for keto-reductase activity against the representative ketone substrate panel.
A seed culture for each strain was grown under appropriate conditions for 16-72 hours in shake flask cultures (250 rpm) containing liquid complex medium (yeasts: yeast extract-peptone-dextrose, 30°C; fungi: Sabouraud dextrose broth + 10 g/L cotton seed flour + 10 g/L yeast extract, 25°C; bacteria: tryptic soy broth, 37°C; and actinomycetes: ISP medium 2, 28°C) to achieve a suitable level of growth. To evaluate biocatalytic activity of “fresh” cultures, seed cultures were harvested by centrifugation and resuspended in fresh complex medium or a defined medium consisting of yeast nitrogen base (with ammonium sulfate) + 20 g/L glucose. Yeast and bacterial cultures were resuspended to an OD600 of 10 while fungi and the actinomycete culture were resuspended in half of the original culture volume. The cell suspensions were then subdivided at 1 mL/well in a 24- or 48-deep well plate. Ketone substrates (Table 2) were added to a concentration of 5 g/L and a “no substrate” control was included. The well plates were sealed with a breathable sealing film and incubated at the appropriate temperature with 250 rpm rotary shaking for 24 hours.
After incubation, 1 mL of acetonitrile was added to each well and the contents were mixed briefly. Following centrifugation to remove cells, the supernatant was analyzed by HPLC. This analytical method consisted of an ODS-Hypersil column (250 x 4.6 mm, 5 μm particle size) and an isocratic mobile phase of 10 mM, pH 2.8 potassium phosphate buffer: acetonitrile (4:1) at a flow rate of 1 mL/min with A210 detection. Percent conversion was calculated as grams of alcohol produced per liter / 5 grams per liter ketone substrate added x 100.
For evaluating biocatalytic activity of cultures after lyophilization, seed cultures were grown as described above and then resuspended in fresh complex or defined medium, each containing 10% (w/v) trehalose as the lyoprotectant. The resuspended cultures were subdivided 1 mL/vial and lyophilized for 24 hours followed by storage at 4°C. Lyophilized cultures were reconstituted with 1 mL of water and transferred to a deep-well plate followed by substrate addition, incubation, sample preparation, and HPLC analysis as described above.
Download a PDF of this application note
Download NowReferences
- Rosenthal K, Lütz S. Recent developments and challenges of biocatalytic processes in the pharmaceutical industry. Curr Opin Green Sustain Chem 11: 58-64, 2018.
- Pollard DJ, Woodley, JM. Biocatalysis for pharmaceutical intermediates: the future is now. Trends Biotechnol 25(2): 66-73, 2007. PubMed: 17184862.
- Hughes DL. Biocatalysis in drug development -- highlights of the recent patent literature. Org Process Res Dev 22(9): 1063-1080, 2018.
- Rodríguez S. Highly stereoselective reagents for beta-keto ester reductions by genetic engineering of baker’s yeast. J Am Chem Soc 123(8): 1547-1555, 2001. PubMed: 11456752.
- Moore, JC, et al. Advances in the enzymatic reduction of ketones. Acc Chem Res 40(12): 1412-1419, 2007. PubMed: 18052114.
- Ferrara MA, et al. Bioconversion of R-(+)-limonene to perillic acid by the yeast Yarrowia lipolytica. Braz J Microbiol 44(4): 1075-1080, 2014. PubMed: 24688495.
- Fan LL, Li HJ, Chen QH. Applications and mechanisms of ionic liquids in whole-cell biotransformation. Int J Mol Sci 15(7): 12196-12216, 2014. PubMed: 25007820.
- Pscheidt B, Glieder A. Yeast cell factories for fi ne chemical and API production. Microb Cell Fact 7: 25, 2008. PubMed: 18684335.
- Sudhakara S, Chadha A. A carbonyl reductase from Candida parapsilosis ATCC 7330: substrate selectivity and enantiospecifi city. Org Biomol Chem 15(19): 4165-4171, 2017. PubMed: 28440822.
- Nakamura K, Matsuda T. Asymmetric reduction of ketones by the acetone powder of Geotrichum candidum. J Org Chem 63(24): 8957-8964, 1998.
- Shah SA, et al. Microbial-catalyzed biotransformation of multifunctional triterpenoids derived from phytonutrients. Int J Mol Sci 15(7): 12027-12060. PubMed: 25003642.
- Sun Z, et al. Metabolic engineering of the L-phenylalanine pathway in Escherichia coli for the production of S- or R-mandelic acid. Microb Cell Fact 10: 71, 2011. PubMed: 21910908.
- Patel RN. Biocatalytic synthesis of chiral alcohols and amino acids for development of pharmaceuticals. Biomolecules 3(4): 741-777, 2013. PubMed: 24970190.
- Wang Y, et al. Engineering of cofactor regeneration enhances (2S,3S)-2,3-butanediol production from diacetyl. Sci Rep 3: 2643, 2013. PubMed: 24025762.
- Katzberg M, et al. Engineering cofactor preference of ketone reducing biocatalysts: a mutagenesis study on a y-diketone reductase from the yeast Saccharomyces cerevisiae serving as an example. Int J Mol Sci 11(4): 1735-1758, 2010. PubMed: 20480039.
- Li Y, et al. Polyketide intermediate mimics as probes for revealing cryptic stereochemistry of ketoreductase domains. ACS Chem Biol 9(12): 2914-2922, 2014. PubMed: 25299319.
- Prakash O, Nimonkar Y, Shouche YS. Practice and prospects of microbial preservation. FEMS Microbiol Lett 339(1): 1-9, 2013. PubMed: 23083094.
- Sigrist CJ, et al. New and continuing developments at PROSITE. Nucleic Acids Res 41(Database issue): D344-D347, 2013. PubMed: 23161676.
- El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res 47(D1): D427-D432, 2019. PubMed: 30357350.
- Wang Q, et al. Characterization and site-directed mutation of a novel aldo-keto reductase from Lodderomyces elongisporus NRRL YB-4239 with high production rate of ethyl (R)-4-chloro-3-hydroxybutanoate. J Ind Microbiol Biotechnol 41(11): 1609-1616, 2014. PubMed: 25189809.