Authors: Mariette Barbier, PhD; and Dev Mittar, PhD
Introduction
Bacterial Detection and Quantification
GFP Expression Does Not Affect Bacterial Growth and Fitness
Fluorescence Detection and Stability
Detection of ATCC GFP-labeled Microorganisms by Fluorescence Microscopy
In Vitro Detection of Host-pathogen Interaction
In Vivo Detection of GFP-labeled P. aeruginosa
Conclusion
Acknowledgments
References
Abstract
We have developed green fluorescent protein (GFP)-labeled strains of various gram-negative opportunistic pathogens from the ATCC collection. These products are compatible with various fluorescence detection technologies such as a fluorescence plate reader, microscopy, flow cytometry, and in vivo imaging systems. The Pseudomonas-GFP pathogenesis model system presented in this application note demonstrates the use of these labeled strains in studying bacterial pathogenesis, host-pathogen interactions, and compound screening assays.
Download a PDF of this application note
Download NowIntroduction
GFP, isolated more than 50 years ago from the jellyfish Aequorea victoria,1 has been used extensively in protein function and localization studies, the analysis of promoter activity and regulation, drug discovery research, and non-invasive imaging.2,3 While GFP has been frequently used in eukaryotic systems, its applications have been limited in microorganisms due to a lack of broad-range molecular tools.
ATCC has developed a vector to express GFP in bacteria for use in pathogenesis and host-pathogen studies. A shuttle vector encoding the GFP variant mut34 (pUCP18-MCSgfpmut3) was generated and successfully transformed into the following ATCC gram-negative opportunistic pathogens (Table 1): Escherichia coli (ATCC 25922), Salmonella enterica subsp. enterica serovar Typhimurium (ATCC 14028), Shigella flexneri (ATCC 12022), and Pseudomonas aeruginosa (ATCC 10145 and ATCC 15692). These GFP strains are compatible with various fluorescence detection technologies, including a fluorescence plate reader, microscopy, flow cytometry, and in vivo imaging systems. GFP-labeled P. aeruginosa was used as a model to test the characteristics of the vector and the sensitivity of detection using these technologies.
Table 1. ATCC GFP-labeled microorganisms
| ATCC number | Species | Reporter |
|---|---|---|
| 25922GFP | Escherichia coli | GFP |
| 14028GFP | Salmonella enterica subsp. enterica serovar Typhimurium | GFP |
| 12022GFP | Shigella flexneri | GFP |
| 10145GFP | Pseudomonas aeruginosa | GFP |
| 15692GFP | Pseudomonas aeruginosa | GFP |
Bacterial detection and quantification
ATCC GFP-labeled microorganisms can be used for a wide range of applications. The expression of a bright GFP variant (gfpmut3) on a high-copy number plasmid facilitates visual identification when exposed to UV-light (Figure 1A) or imaged using a detection system such as the IVIS Spectrum (PerkinElmer) (Figure 1B and Figure 1C). Quick colony differentiation of GFP-labeled microorganisms from unlabeled organisms or contaminants can also be easily performed by using a hand-held UV wand.
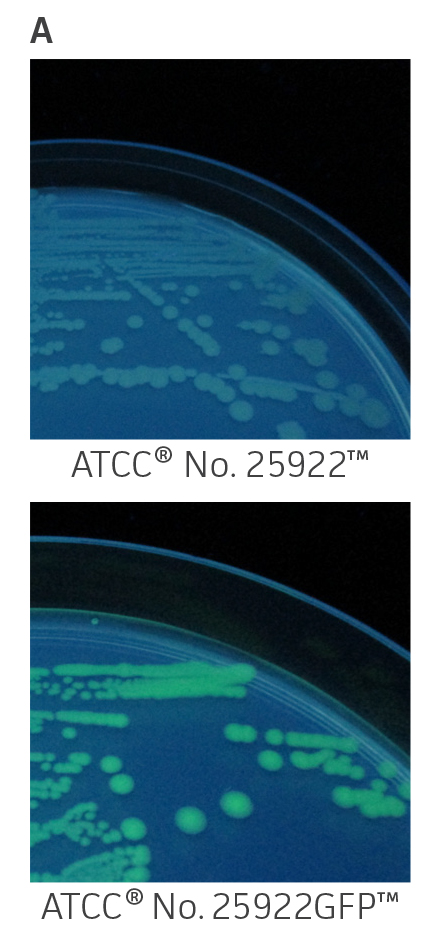
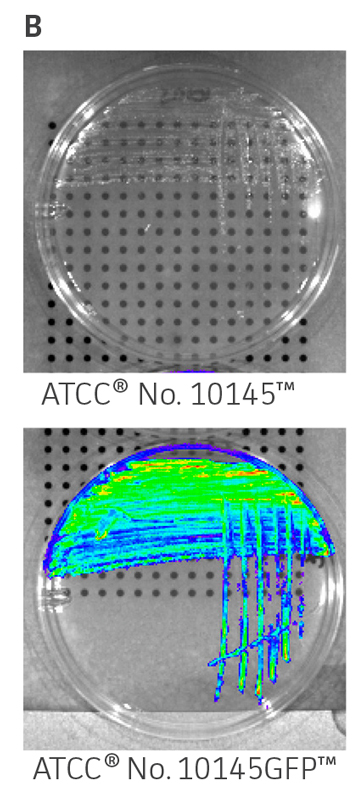
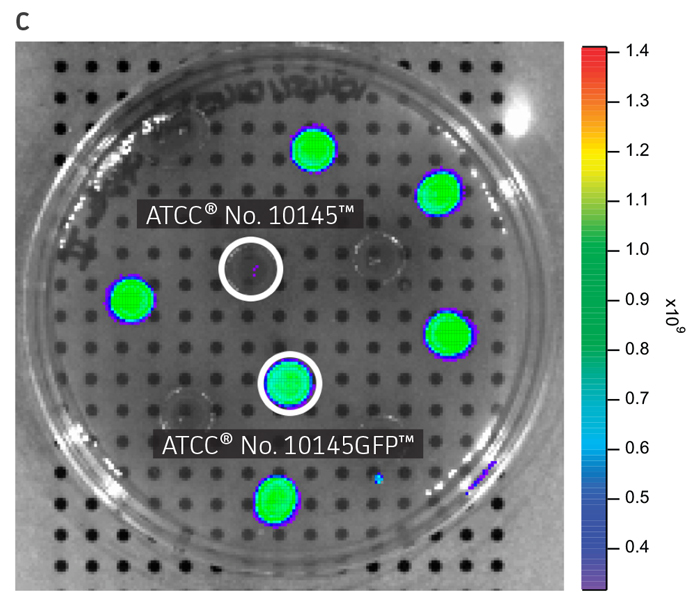
Figure 1. Visual detection of GFP-labeled microorganisms. (A) An empty vector control strain (ATCC 25922) and E. coli-GFP (ATCC 25922GFP) were grown at 37°C in Lysogeny agar (LA) with 100 μg/mL carbenicillin. Colonies were imaged using a UV Wand (Ultra Violet Products). (B) An empty vector control strain (ATCC 10145) and P. aeruginosa-GFP (ATCC 10145GFP) were streaked on LA with 300 μg/mL carbenicillin, incubated for 24 hours (h) at 37°C, and imaged using an IVIS Spectrum detection system (Ex. = 500 nm, Em. = 540 nm). (C) Mixture of P. aeruginosa (ATCC 10145) and P. aeruginosa-GFP (ATCC 10145GFP) colonies on LA with 300 μg/mL carbenicillin, incubated for 24h at 37°C, and imaged using an IVIS Spectrum detection system (Ex. = 500 nm, Em. = 540 nm). Key to fluorescence intensity units is shown on the right.
GFP expression does not affect bacterial growth and fitness
To determine whether GFPmut3 expression affects growth fitness and viability of the labeled microorganisms (Table 1), the expression of the gfpmut3 gene was monitored during growth (Figure 2). GFPmut3 labeled P. aeruginosa showed the same growth profiles as the parental strain carrying the empty vector (Figure 2A). Fluorescence was easily detected and quantified using a fluorescence microplate reader (Figure 2B). Background fluorescence of the non-labeled wild-type strain was extremely low. A linear correlation was observed between fluorescence and colony forming units (cfu) (R2=0.998) or optical density (R2=0.999) at concentrations ranging from 106 to 1010 CFU/mL (Figure 2C). These findings show that GFPmut3 labeling does not affect bacterial growth and can easily be used to detect and quantify bacteria using a fluorescence microplate reader.
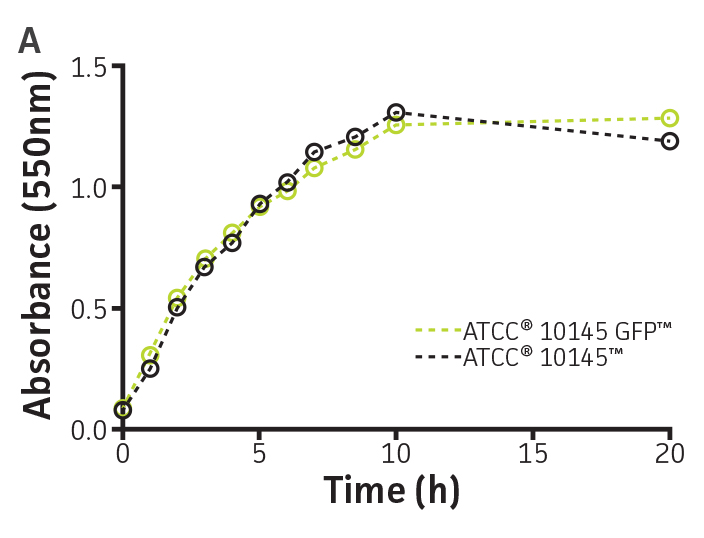
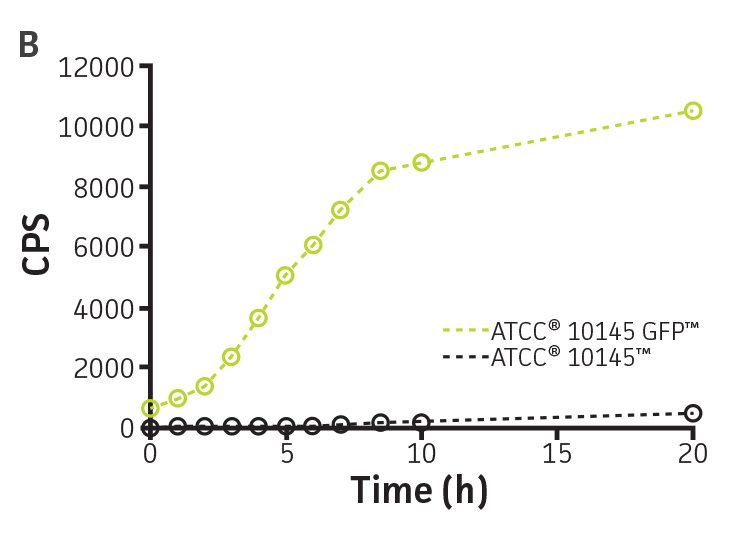
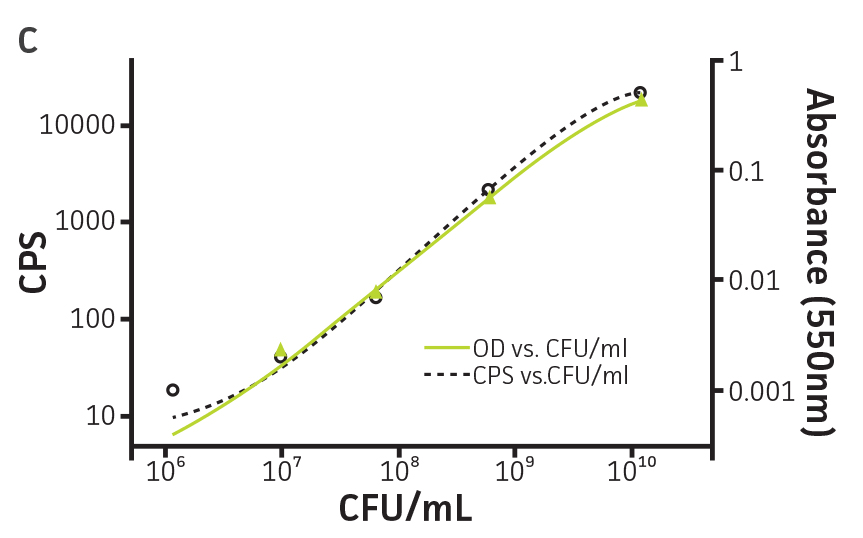
Figure 2. Expression of GFPmut3 does not affect bacterial growth and can be used for bacterial quantification. P. aeruginosa-GFP (ATCC 10145GFP) (green) and an empty vector control (ATCC 10145) (black) were grown at 37°C in Lysogeny broth (LB) with 300 μg/mL carbenicillin under constant shaking. (A) Absorbance at 550 nm and (B) fluorescence expressed as counts per second (CPS) (Ex. = 490 nm, Em. = 535 nm) were determined at regular intervals (VICTOR X3 Multilabel Plate Reader, PerkinElmer). (C) Colony forming units (CFU) were determined by plating appropriate dilutions on LA plates and were compared to CPS (black, R2=1.000) and absorbance (green, R2=0.998) using a non-linear regression.
Fluorescence detection and stability
The stability of the plasmid was tested in P. aeruginosa-GFP by passaging a culture in the absence of antibiotic pressure. This was analyzed by measuring fluorescence (Ex. = 490 nm, Em. = 535 nm) every 24h on a microplate reader (Figure 3A and Figure 3B). The plasmid was stable in P. aeruginosa-GFP (ATCC 10145GFP) for 3 successive passages in the absence of antibiotics and for a minimum of 20 generations (Figure 3C). Together with the long half-life of GFPmut3 in bacteria (over 3 days, data not shown), these results indicate that the fluorescence of GFPmut3 labeled microorganisms is very stable.
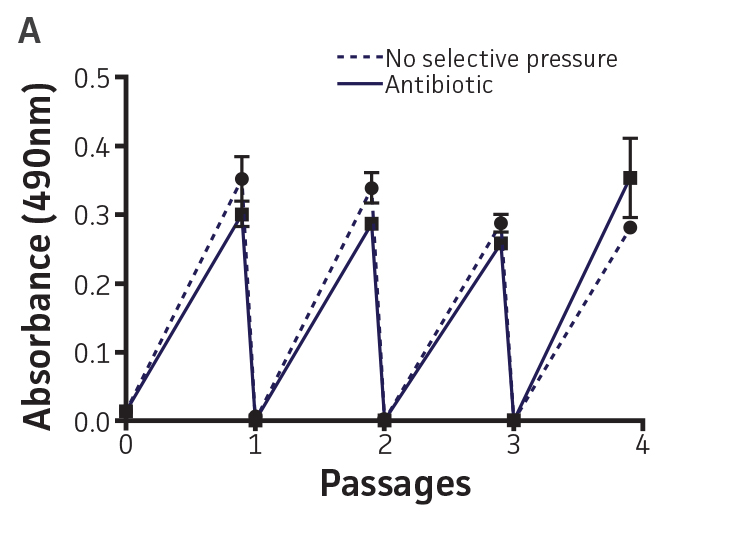
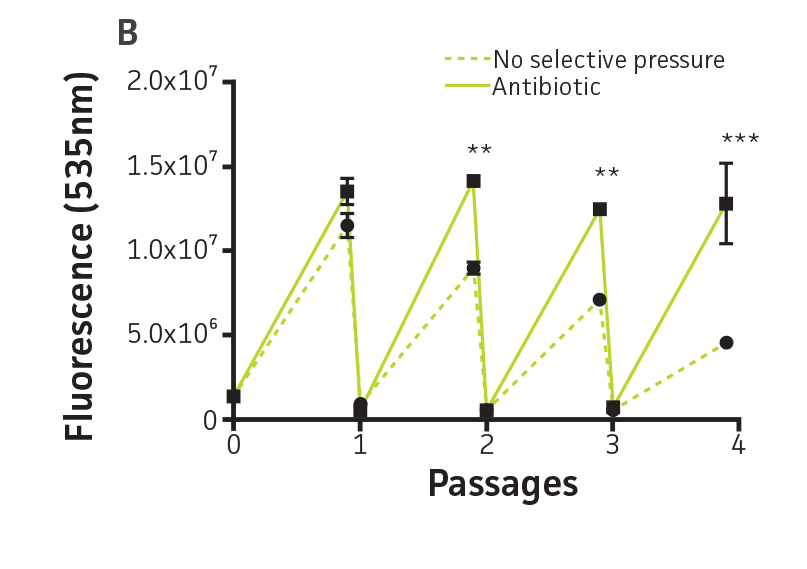
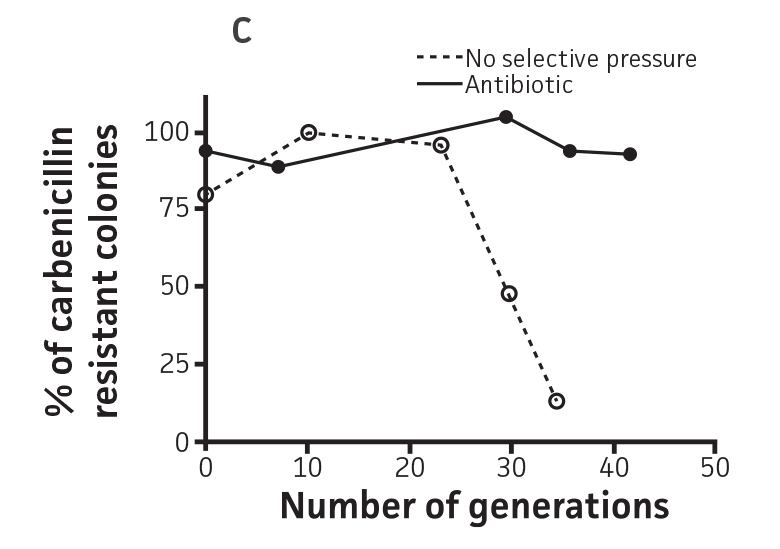
Figure 3. Plasmid stability in P. aeruginosa-GFP (ATCC 10145GFP). P. aeruginosa-GFP (ATCC 10145GFP) was grown at 37°C in LB with 300 μg/mL carbenicillin (solid line), or in the absence of antibiotic pressure (dashed line), under constant shaking. Cultures were passaged every 24h and diluted 100 times in fresh medium. (A) Absorbance at 490 nm, and (B) fluorescence (Ex. = 490 nm, Em. = 535 nm) were determined at regular intervals on a microplate reader. (C) Generation number was determined by plating cultures on selective and non-selective LA plates. Data were analyzed with an unpaired two-tailed t-test, and significant data are denoted with an asterisk (**: p<0.001; ***: p<0.0001).
Detection of ATCC GFP-labeled microorganisms by fluorescence microscopy
The utility of fluorescent proteins is often limited by their sensitivity to photo bleaching. The resistance of P. aeruginosa-GFP (ATCC 10145GFP) to photo bleaching was tested under continuous exposure to UV light. The fluorescence remained stable during the first 5 minutes (m), a significant loss of signal was however observed after 7m of exposure to UV light. The GFPmut3 reporter used in this study therefore shows resistance to photo bleaching in P. aeruginosa (Figures 4A and Figure 4B).
To visualize bacterial interaction with eukaryotic cells, P. aeruginosa-GFP (ATCC 10145GFP) was incubated with A549 (ATCC CCL-185) airway epithelial cells followed by fixation with 2% paraformladehyde and counterstaining of nuclei (blue) and actin (red). The pseudo-colored images show that fluorescently labeled P. aeruginosa-GFP (ATCC 10145GFP) can be detected and used for studying interactions with mammalian cells by fluorescence microscopy (Figure 4C).
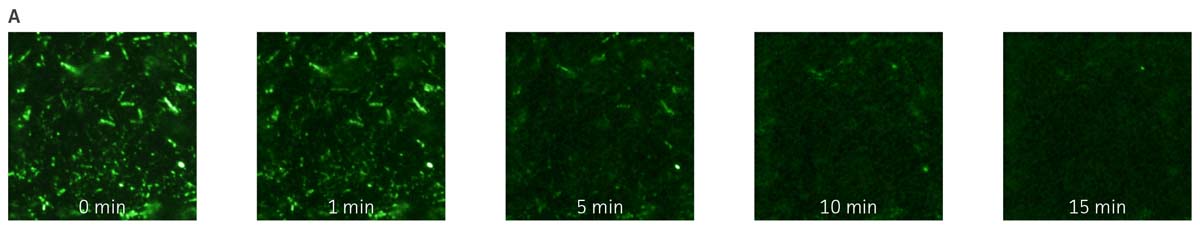
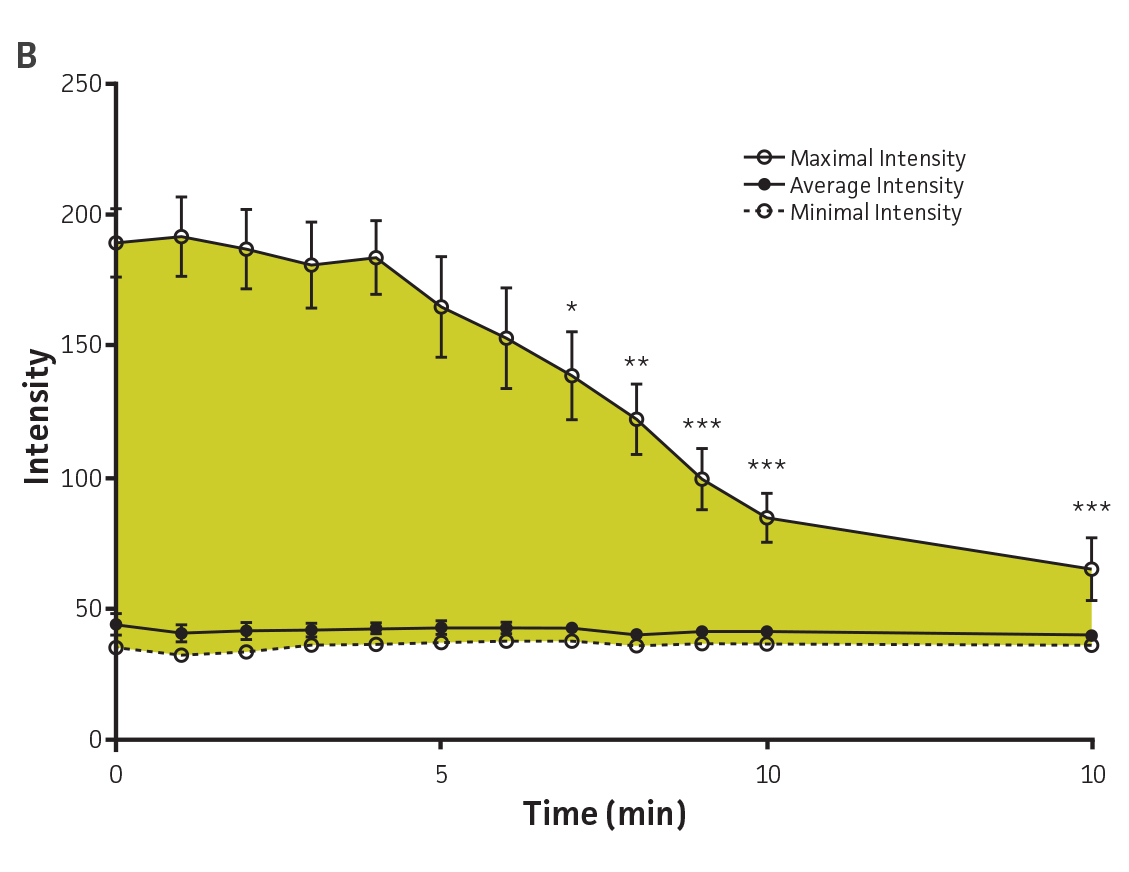
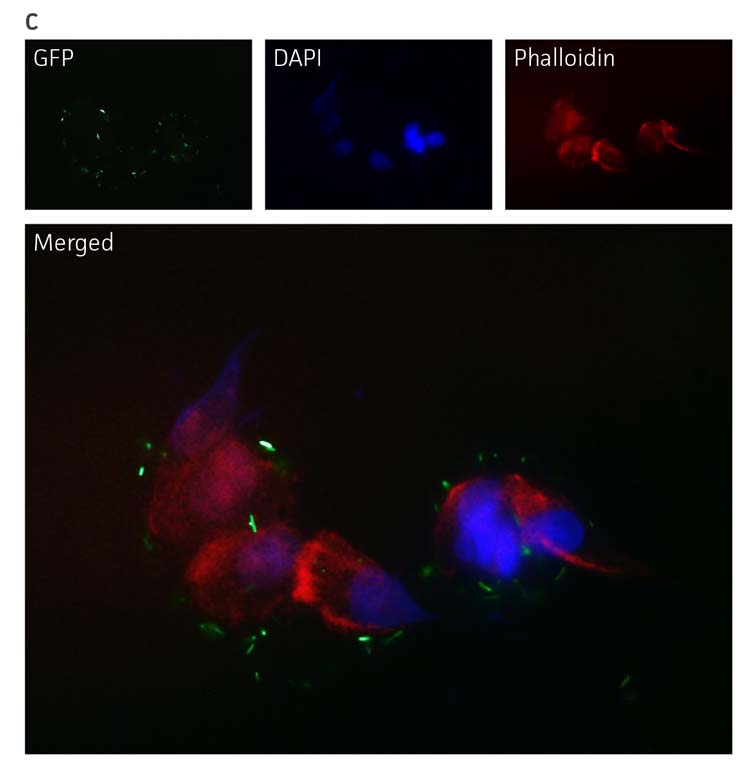
Figure 4. Detection of P. aeruginosa-GFP (ATCC 10145GFP) by fluorescence microscopy. (A) P. aeruginosa-GFP (ATCC 10145GFP) was exposed to UV light (100W) and imaged every 60 seconds (Axioskop, ZEISS, × 100 oil immersion, 505 nm filter). (B) Image intensity was measured in 9 independent areas of 200 × 200 pixels using Image J (rsbweb.nih.gov/ij/), and is represented in arbitrary units. Data were analyzed with an unpaired two-tailed t-test and data significant from the non exposed control are denoted by an asterisk (*: p<0.01; **: p<0.001; ***: p<0.0001). (C) P. aeruginosa-GFP (ATCC 10145GFP) interaction with A549 (ATCC CCL-185) airway epithelial cells (×100 oil immersion). Bacteria appear in green (505 nm), nuclei were stained with DAPI (475 nm, blue) (Invitrogen), and the cytoskeleton was stained with Alexa Fluor 555 Phalloidin (565 nm, red) (Invitrogen).
In vitro detection of host-pathogen interaction
P. aeruginosa-GFP (ATCC 10145GFP) was tested in a high-throughput adhesion and internalization assay in 96-well plates using a fluorescence microplate reader and by flow cytometry. The sensitivity of the assay was tested using a phagocytosis inhibitor (Cytochalasin D, Sigma-Aldrich). Bacterial attachment and internalization by airway epithelial cells was detected using a fluorescence microplate reader (Figure 5A). This proof-of-concept experiment indicates that these fluorescent microorganisms can be used in high-content in vitro assays in 96-well plates to screen small molecule inhibitors and also to study pathogen-host interactions.
Bacterial fluorescence (Figure 5B), attachment, and internalization by murine macrophages (Figure 5C) were also detected using a flow cytometer. These data show that macrophages with GFP-labeled Pseudomonas can be detected using flow cytometry. Overall, GFPmut3 labeled reporters can be used for studying interactions of various pathogens with eukaryotic cells using flow cytometry.
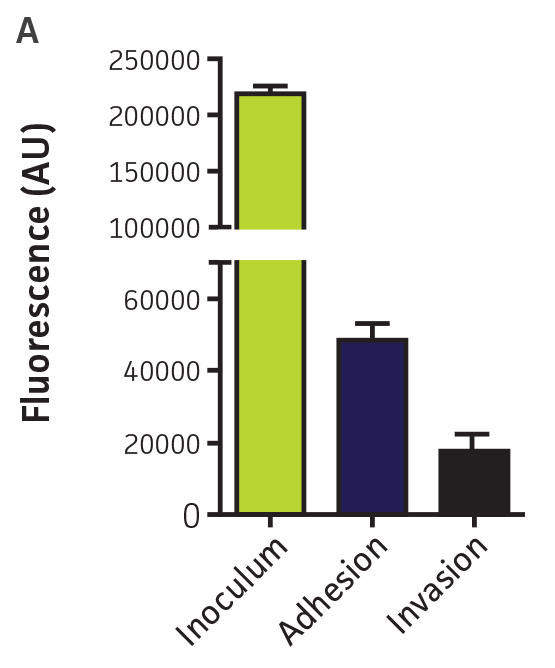
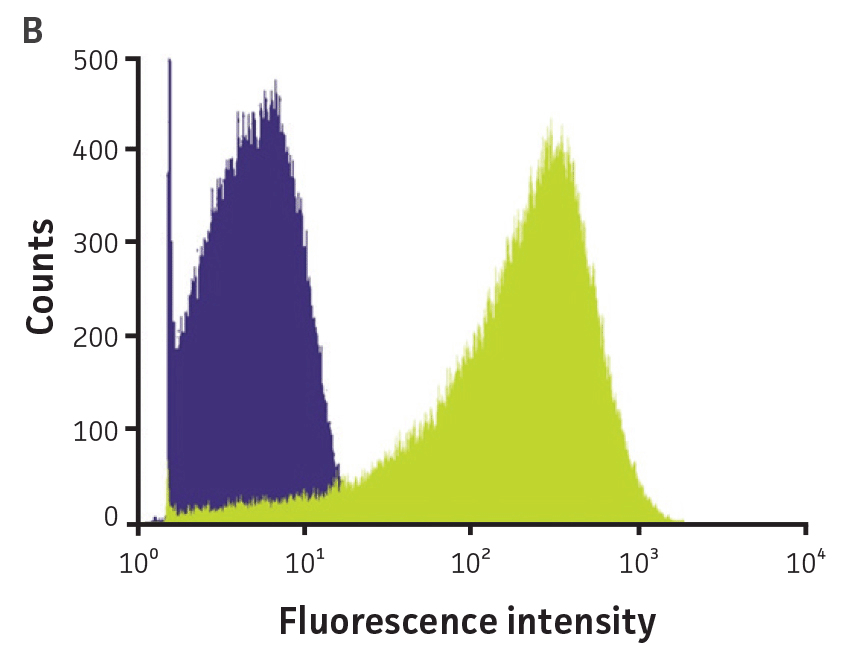
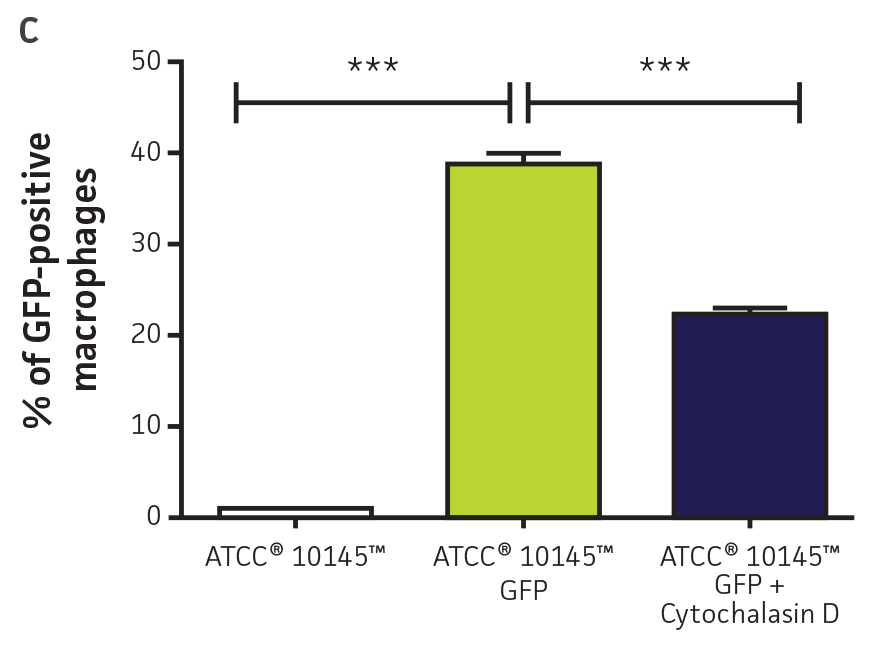
Figure 5. High-throughput detection of pathogen-host interactions. (A) P. aeruginosa-GFP (ATCC 10145GFP) was incubated in the presence of A549 (ATCC CCL-185) airway epithelial cell monolayers in 96-well plates at a multiplicity of infection (MOI) of 50. After 1h, the cells were washed (Adhesion) and the medium was replaced by F12-K supplemented with 100 μg/mL of gentamicin for 1h to kill extracellular bacteria. Cells were washed again and fluorescence was measured in arbitrary units (AU) on a microplate reader (Invasion) (Ex. = 490 nm, Em. = 535 nm). (B) P. aeruginosa (ATCC 10145) (purple) and P. aeruginosa-GFP (ATCC 10145GFP) (green) bacterial suspensions were analyzed by flow cytometry using a FACSCalibur (BD). (C) P. aeruginosa (ATCC 10145) and P. aeruginosa-GFP (ATCC 10145GFP) were incubated with J774A.1 (ATCC TIB-67) macrophages at a MOI of 20 in the presence or absence of 50 nM Cytochalasin D and analyzed by flow cytometry. Data were analyzed with an unpaired two-tailed t-test, and significant values are denoted with an asterisk (***: p<0.0001).
In vivo detection of GFP-labeled P. aeruginosa
To test the utility of this GFP-reporter for pathogenesis studies, we used an established plant host model system5 that has been successfully used for high-throughput analysis of virulence and in drug discovery. The mid-ribs of Lactuca sativa L. var. longifolia were infected with various concentrations of P. aeruginosa-GFP (ATCC 10145GFP), and fluorescence was measured using an in vivo imaging system. The assay detected 106 to 109 CFU/site of infection, indicating that this vector can be successfully employed to monitor bacterial growth in the plant host (Figure 6).
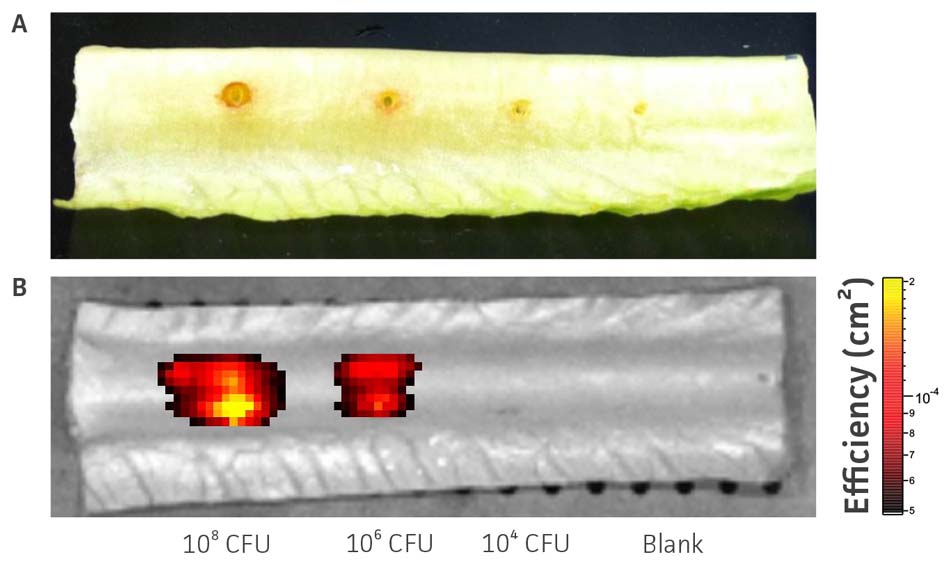
Figure 6. In vivo detection of P aeruginosa-GFP (ATCC 10145GFP). Various doses of P. aeruginosa-GFP (ATCC 10145GFP) were injected into the mid-rib of Lactuca sativa L. var. longifolia. (A) The area of necrosis caused by P. aeruginosa-GFP (ATCC 10145GFP) was measured in mm. (B) Fluorescence was detected after 48h using an IVIS Spectrum detection system (Ex. = 500 nm, Em. = 540 nm).
Conclusion
ATCC has developed a vector to express GFP in pathogenic bacteria for use in bacterial pathogenesis and host-pathogen interaction studies. The vector is stable and GFP expression did not alter bacterial growth. The GFP-labeled P. aeruginosa (ATCC 10145GFP) model system represents a proof-of-concept for detecting gram-negative bacteria using a fluorescence plate reader, microscopy, flow cytometry, and in vivo imaging technologies. Similarly, P. aeruginosa (ATCC 15692GFP), E. coli (ATCC 25922GFP), S. enterica subsp. enterica serovar Typhimurium (ATCC 14028GFP), and S. flexneri (ATCC 12022GFP) can be used for similar research and compound screening applications using a multitude of detection systems.
Download a PDF of this application note
Download NowAcknowledgments
The authors would like to thank Dr. Erik L. Hewlett and Dr. F. Heath Damron from the University of Virginia for their help on this project.
References
- Shimomura, O., Johnson, F. H. & Saiga, Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59, 223-239 (1962).
- O’Toole, R., Von Hofsten, J., Rosqvist, R., Olsson, P. E. & Wolf-Watz, H. Visualisation of zebrafish infection by GFP-labelled Vibrio anguillarum. Microb Pathog 37, 41-46, doi:10.1016/j.micpath.2004.03.001 (2004).
- Stuurman, N. et al. Use of green fluorescent protein color variants expressed on stable broad-host-range vectors to visualize rhizobia interacting with plants. Mol Plant Microbe Interact: MPMI 13, 1163-1169, doi:10.1094/MPMI.2000.13.11.1163 (2000).
- Choi, K. H. & Schweizer, H. P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1, 153-161, doi:10.1038/nprot.2006.24 (2006).
- Starkey, M. & Rahme, L. G. Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat Protoc 4, 117-124, doi:10.1038/nprot.2008.224 (2009).