Why it's important
The HCMI portfolio is an incredibly diverse collection derived from primary patient tumor samples from clinical sites across the US and Europe. The collection includes over 20 cancer types from hundreds of unique human donors, more than 25 media formulations, and models that are maintained in a variety of different culture conditions, both 2-D and 3-D. In addition to deriving the model, the HCMI provides a searchable catalog and annotates the models with molecular characterization via sequencing and donor clinical data that is made available from the NCI at no cost. This allows researchers to identify models with specific genotypes of interest, such as specific mutations or combinations of mutations or specific patient demographics and treatment histories.
The initiative holds promise to greatly expand the breadth, depth, physiological relevance, and level of molecular characterization of in vitro cancer models available to the global research community. For example, with the inclusion of HCMI models, ATCC’s portfolio of glioblastoma models has roughly doubled, and nearly half of the new models are 3-D models, which may better reflect in vivo physiology than existing cell lines. The number of cancer organoid models available from ATCC has increased from zero three years ago to over 130 today, enabling this new 3-D culture technology to be easily accessible to laboratories that don’t have the ability to derive their own patient-derived cancer models.
What we do
ATCC is the sole distributor for the HCMI models, and we work closely with the NCI and model developers to support the goal of ensuring these models are broadly available to the research community. Beyond manufacturing and distribution, we also perform critical authentication and quality control activities to ensure the models satisfy ATCC’s high quality standards. We also strive to provide customers with educational materials and tools for facilitating use of these models. Specifically for 3-D cancer organoids, we published a protocol paper detailing how they can be thawed into culture, expanded, and cryopreserved, and we also released an organoid culture video tutorial.
ATCC is excited to collaborate with the NCI and will continue supporting scientists in using these more complex models. New models will continue to be released as well as tools for easier adoption of organoids.
James Clinton, PhD
Lead Scientist, ATCC
James Clinton, PhD, works in new product development with a focus on primary cells and advanced, physiologically relevant culture systems using novel technologies. Previously he worked at University of California, San Diego and the La Jolla Institute for Molecular Medicine. Dr. Clinton attended Washington State University and University of California, San Diego where he studied Neuroscience.
Dig into the HCMI
Human Cancer Models Initiative (HCMI)
ATCC is the exclusive distributor of the Human Cancer Models Initiative (HCMI) models. See the models that include common and rare examples of cancer from numerous tissues.
More Culture guide
Culture guide
Organoid Culture Guide
Get expert guidance on growing and maintaining organoids from our organoid culture guide.
More Webinar
Webinar
Transforming In Vitro Cancer Research With Next-generation Biological Models from the Human Cancer Models Initiative
There is an unmet need for preclinical cancer models that better reflect the genotype and phenotype across the spectrum of cancer found in the patient population. Watch this webinar to hear about the wide variety of patient-derived in vitro cancer models ATCC offers in collaboration with HCMI, including organoids, neurospheres, and conditionally reprogrammed cells.
More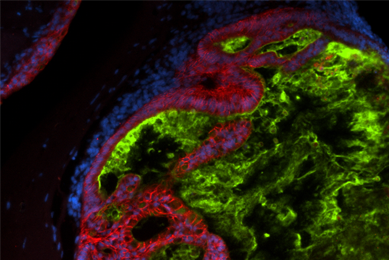
Tutorial: Thawing, Culturing, and Cryopreserving Human Organoids
Watch a video to learn how to culture organoids.
More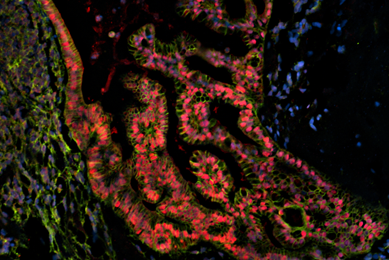
Organoids
Patient-derived organoids are authenticated cell models paired with genomic and phenotypic data. Organoids are available from the Human Cancer Models Initiative (HCMI) and contribute to valuable and reproducible research.
More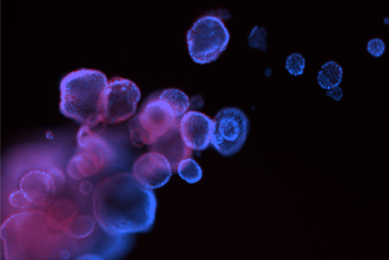
Cell Models
Cell models are widely used in toxicology, immunology, and cancer pharmacogenomic studies to predict clinical response and to identify novel mechanisms associated with variation in drug response.
More