This guide contains general technical information for bacterial growth, propagation, preservation, and application. Additional information on bacterial culturing can be found in Bergey’s Manual of Systematic Bacteriology, 2nd Edition, published by Springer, New York.1
Table of Contents
Staying Safe in a Pandemic Environment
Getting Started with an ATCC Bacterial Strain
Bacterial Growth and Propagation
Growth Media
Preservation
Biosafety and Disposal
Bacterial Authentication
Bacterial Applications
Glossary
References
Get a printer-friendly copy of the Bacteriology Culture Guide to keep your research on track
Download NowStaying Safe in a Pandemic Environment
When the recent coronavirus pandemic hit, laboratories throughout the world resolved to shut down operations, reduce the scale of work, or proceed at full steam. To safeguard the health of our scientists, ATCC has adopted a battery of best practices that minimize transmission of SARS-CoV-2 with little impact on productivity. Whether returning after a hiatus or gearing up for a new project, we can all use a refresher to help follow best practices. Please read this first section of the culture guide for some quick reminders about common contamination hotspots and advice on how to keep them in check while getting your work done.
Materials, Contamination, and Workspace
You may be just getting back into the laboratory or beginning a new project. Before you start, consider some potential hotspots that can profoundly affect your experimental results, such as the quality of your starting materials, execution of proper laboratory technique, and organization of your workspace. Here are some simple tips and techniques to avoid ruining your experiments, leading to confounding results, paper retractions, financial loss, and damaged reputation.
Material for Research
- Check existing materials for signs of contamination.
- Authenticate and replenish your cell lines and microbes.
- Start new projects with trustworthy materials.
- Routinely check the expiration dates of media and reagents.
Cross Contamination
- Ensure everyone—new and experienced—is trained on aseptic techniques.
- Aliquot your samples and reagents.
- When aliquoting is impractical, put just the amount of the reagent you expect to use into a secondary container. Discard the remainder when finished working.
- Avoid sharing pipettes or other equipment.
- Clean the insides and exteriors of pipettes and tools that must be shared.
- Utilize the biosafety cabinet to reduce contamination.
Personal Space and Equipment
- Keep bench space uncluttered.
- Wash your lab coat regularly.
- Leave personal items outside.
- Clean your work area before and after use.
- Use lab tablets instead of personal phones.
- If personal items are needed, sanitize them before and after lab use.
- Move extra equipment away from walls and crevices to facilitate frequent and thorough cleaning.
Avoiding Infection
Like you, we’re committed to protecting the health of our colleagues. While SARS-CoV-2 is currently unique in its pathogenic nature and transmission dynamics, other infectious organisms may in time arise to threaten the health of laboratory workers. To reduce the chance of contracting a current or emerging infectious disease while working in the lab under epidemic or pandemic conditions, we recommend you follow these best practices.
Coming and Going
- Wash your hands well when entering and leaving the lab.
- Master the basics of proper personal protective equipment (PPE) use and removal.
- Stay home if you’ve been exposed to any illness.
- Inspect PPE prior to use
Interacting With Others
- Remember, particles spread via talking, coughing, and breathing.
- Use virtual collaboration tools, and only converse before or after working.
- Always keep your nose, mouth, and skin covered with PPE.
- Be extra vigilant about PPE use when working with animals.
- Keep 6 feet of space between individuals.
Airborne Transmission
- Reduce foot traffic in the lab.
- Designate one-way traffic flows to support distancing.
- Try limiting capacity to aid physical distancing.

Getting Started with an ATCC Bacterial Strain
ATCC bacterial strains are shipped frozen on dry ice in plastic cryopreservation vials, as lyophilized cultures in glass ampoules or serum vials, or as live cultures on agar slants or in broth medium. Upon receipt of frozen cultures, immediately revive cultures by thawing and subsequently transferring cultures to an appropriate growth medium. If this is not possible, store frozen vials in liquid nitrogen vapor (below -130°C). Alternatively, the frozen material can be stored between -70°C and -80°C for short periods (1 to 5 days); however, viability will decline at temperatures above -130°C. Freeze-dried cultures can be stored safely at 4°C or lower. Upon receipt, rehydrate or dilute cultures using the appropriate medium and incubation conditions specified for the product. Live cultures should be transferred to an appropriate growth medium upon receipt and incubated under the conditions specified for the product.
Product Sheet
ATCC bacterial strains typically have detailed information on the initiation and expansion of materials as well as ideal growth and propagation conditions. The complete information for a product can be found on the product detail page for that specific material. In addition, a product sheet that contains curated information for safety and handling can be found on atcc.org.
Nomenclature Changes
Changes in taxonomy or further analysis of bacterial strains may lead to a change in nomenclature. Further information on bacterial nomenclature changes can be found on the List of Prokaryotic Names with Standing in Nomenclature (LPSN), which can be accessed online at www.bacterio.net/.
Preparation of Medium
In advance, prepare the appropriate medium and additional reagents necessary for bacterial strain revival and growth. Ensure that the proper incubation conditions are ready. Information for the formulation and preparation of the media and incubation conditions for these products is available on the product detail page for the product on atcc.org.
Opening Glass Ampoules
Overview
All cultures should be considered potentially hazardous and should be opened by individuals trained in microbiological techniques working in facilities with containment requirements appropriate for the biosafety level of the cultures. ATCC recommends that the handling or opening of glass ampoules be performed in a biological safety cabinet. If this is not possible, wear protective clothing, gloves, a face shield or safety goggles, and hold the vial away from your body. Ensure that all empty vials are sterilized before disposal.
Opening Double-Vial Preparations (see diagram)
- Heat the tip of the outer vial in a flame.
- Add a few drops of water on the hot tip to crack the glass.
- Strike the end of the vial with a file or pencil to remove the tip.
- Remove the insulation and inner vial with sterile forceps. Gently raise the cotton plug.
Opening Single-Vial Preparations (see diagram)
- To recover the cell suspension from the glass ampoule, score the neck of the ampoule with a small, sterile file.
- Disinfect the outside of the ampoule with freshly prepared 70% ethanol or dip it into a beaker of freshly prepared 70% ethanol.
- Wrap the ampoule within several folds of a sterile towel or gauze to dry residual ethanol.
- Working in a laminar flow hood, hold the vial upright and snap open the vial. Ensure that your gauze does not become too wet with ethanol, or alcohol could be sucked into the culture when the vacuum is broken. Rehydrate the material immediately.
Initiating Frozen Cultures
- Prepare a sterile test tube that contains the recommended medium for bacterial growth as specified for the product. Ensure that the medium contains all necessary reagents and is equilibrated for temperature and pH.
- Thaw the sample vial via gentle agitation in a water bath that is set to the normal growth temperature of that strain. Thawing will be rapid; approximately 2 minutes or until all ice crystals have melted.
- Remove the vial from the water bath and decontaminate the outer surface using 70% ethanol. Follow strict aseptic conditions in a laminar flow hood for all further manipulations.
- Unscrew the top of the vial and transfer the entire contents to a sterile test tube containing the appropriate growth medium. Additional test tubes can be inoculated by transferring 0.5 mL of the primary culture to additional secondary cultures.
- Incubate cultures under the appropriate temperature and atmospheric conditions as recommended for the product.
- Examine cultures after the recommended incubation period. The incubation period will vary between strains. (See: NOTE 1)
NOTE 1
Following recovery from cryopreservation or lyophilization, some bacterial strains may exhibit a prolonged lag phase. These strains may require an extended incubation period.
Initiating Lyophilized Cultures
- Using a Pasteur pipette, aseptically add 0.5 mL of the recommended growth medium to the freeze-dried material. Mix well.
- Transfer the entire suspension to a test tube containing 5 to 6 mL of the recommended medium. Additionally, transfer several drops of the suspension to an agar slant.
- Incubate cultures under the appropriate temperature and atmospheric conditions as recommended for the product. Additional test tubes can be inoculated by transferring 0.5 mL of the primary culture to additional secondary cultures.
- Examine cultures after the recommended incubation period. The incubation period will vary between strains. Most freeze-dried cultures will grow within a few days. (See: NOTE 1)
Handling Test Tube Cultures
- Incubate the culture upon receipt under the appropriate temperature and atmospheric conditions recommended for the product. Do not store the culture in a refrigerator.
- Transfer the culture to fresh media as specified for the product. When transferring a broth culture, aseptically withdraw approximately 1.0 mL of the culture and transfer into 5 mL of fresh broth, or transfer several drops of the suspension to an agar slant or plate. When transferring a culture from an agar slant, aseptically transfer a single colony to 5 mL of fresh broth or to an agar slant or plate.
- Incubate the culture under the appropriate temperature and atmospheric conditions recommended for the product.
Bacterial Growth and Propagation
Bacterial Growth Conditions
Temperature
Because bacteria can grow and thrive in a variety of environments, optimal growth temperatures may vary significantly between species. In general, most pathogenic or commensal bacterial strains grow well at body temperature (37°C). In contrast, many environmental strains thrive at lower temperatures, often within a range of 25°C to 30°C.
Bacterial species can be categorized based on their growth temperature; these include psychrophiles (0°C to 20°C), mesophiles (25°C to 40°C), and thermophiles (45°C to 122°C). Though bacterial strains require optimal temperatures for growth and reproduction, most strains can withstand considerable drops in temperature and survive several days at 4°C. At these lower temperatures, bacterial growth and metabolism are significantly diminished. (See: NOTE 2)
NOTE 2
Regularly calibrate the temperature control system of incubators. Use an alarm system when possible to warn against temperature increases above the optimum setting.
Atmosphere
In addition to varying requirements for optimal growth temperatures, bacteria also differ in their use of oxygen for respiration. Aerobic organisms, such as Bacillus species, use oxygen as a terminal electron acceptor during respiration. Similarly, microaerophiles, such as Helicobacter pylori, also require the use of oxygen, but at lower levels than naturally occurring in the environment. In contrast, anaerobic organisms use electron acceptors such as nitrate or sulfate, among other inorganic acceptors. These inorganic compounds, however, have a lower reduction potential than oxygen thus resulting in less efficient respiration.
The use of oxygen and inorganic compounds by anaerobic organisms can differ greatly between species. Obligate anaerobes, such as Clostridium species, can only survive and reproduce in the absence of oxygen; these organisms are often killed by the presence of oxygen. Similarly, aerotolerant anaerobes, such as Lactobacillus species, cannot use oxygen during respiration; however, unlike strict anaerobes, these microorganisms can tolerate oxygen for short periods of time. Lastly, facultative anaerobes, such as Escherichia coli and Staphylococcus species, are able to survive in both the presence and absence of oxygen. If given the choice, these organisms prefer the use of oxygen during respiration as it has the greatest reduction potential as compared to other electron acceptors.
When working with anaerobic cultures, it is important to avoid unnecessary exposure to oxygen. Anaerobic conditions may be obtained for either transfer or incubation by the methods listed below. Unless specifically mentioned, the standard anaerobic gas mixture is 80% N2, 10% CO2, and 10% H2.
Anaerobic conditions for transfer may be obtained by either of the following:
- Use of an anaerobic gas chamber.
- Placement of test tubes under a gassing cannula system hooked to anaerobic gas.
During incubation, anaerobic conditions may be maintained by any of the following:
- Loose screw-caps on test tubes in an anaerobic chamber.
- Loose screw-caps on test tubes in an activated anaerobic GasPak system.
- Use of sterile butyl rubber stoppers on test tubes so that an anaerobic gas headspace is retained.
Bacterial Propagation
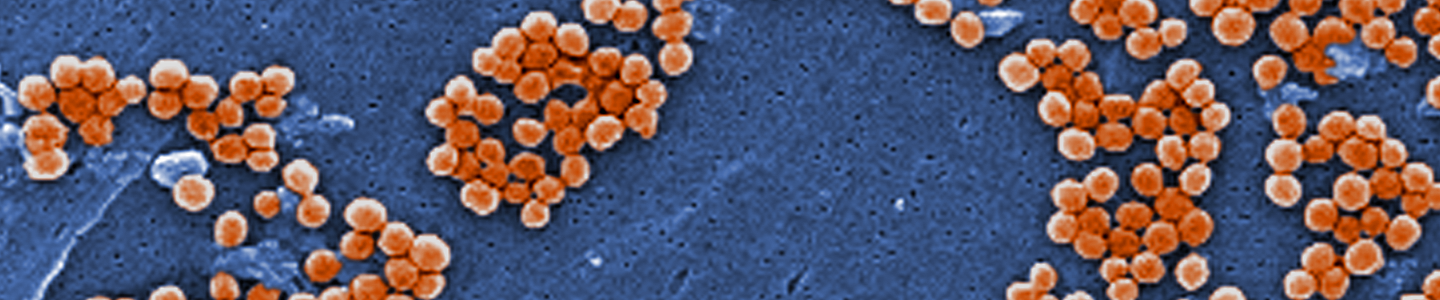
 Image of MRSA courtesy of Annie Cavanagh, Wellcome Images
Image of MRSA courtesy of Annie Cavanagh, Wellcome ImagesPropagation of bacterial strains can vary significantly between species. Below, we describe the general procedure for the propagation of non-fastidious strains and a more detailed set of procedures for fastidious strains and bacteriophages.4 Information regarding the recommended medium and growth conditions can be found on the product detail page. The product sheet, as well as additional information, can be found on the ATCC website or can be requested from ATCC Product Experience. Additional propagation information can also be found in Bergey’s Manual of Systematic Bacteriology 2nd Edition, Published by Springer, New York.1
Propagation of Non-Fastidious Strains
- Open the vial according to enclosed instructions.
- Initiate the lyophilized or frozen culture according to the instructions described in the previous chapter, "Getting Started with an ATCC Bacterial Strain." Be sure to rehydrate the entire pellet for optimal recovery.
- Aseptically transfer this aliquot back into the broth tube. Mix well.
- Use several drops of the suspension to inoculate the recommended agar slant and/or plate.
- Incubate the tubes and plate at the recommended temperature and atmospheric conditions. The incubation period will vary between species.
Propagation of Bacteriophages and Fastidious Strains
- Bacteriophages
ATCC bacteriophages should be propagated in their respective bacterial host strain.
To recover phage from freeze-dried or thawed liquid nitrogen vials: - Prepare an actively growing broth culture of the recommended host strain before opening the phage specimen. The host should be in early log phase.
- Add approximately 0.25 mL of the recommended host broth to a freeze-dried phage.
- Pre-warm plates of the recommended medium in an incubator. Overlay the surface with 2.5 mL of melted 0.5% agar (same medium) that contains one or two drops of the freshly grown host. The soft agar should be maintained at 43°C to 45°C until ready to pour. It may be advisable to use a water bath. Allow the overlay to harden.
- The re-hydrated phage can be serially diluted using a 1:10 dilution scheme by passing 0.25 mL of the phage into a tube containing 2.25 mL of the broth medium for freeze-dried vials; 0.5 mL of the phage into a tube containing 4.5 mL of the broth media for frozen vials. Repeat for as many passages as desired.
- One drop of each dilution is spotted on the surface of the prepared plates. Allow this to dry. Three to four dilutions can be placed on each plate. After 24 hours incubation, lysis should be visible. At the higher dilutions, individual plaques should be countable.
- Many strains may also be titrated without a soft agar overlay. Pipette approximately 1.0 mL of the host onto the surface of each plate. After tilting plate to ensure the entire surface is covered, the excess liquid is aspirated off. After the surface dries, the various dilutions of the phage are dropped onto the surface as before. (See: NOTE 3)
- Phage may be propagated by preparing plates with the soft agar/host overlay as above and covering the surface with approximately 0.5 mL of the concentrated phage. Or, alternatively, you may add the phage directly to the melted agar/host before pouring over the plates. For larger amounts, large-size T flasks can be prepared with the recommended agar, and approximately 12.0 mL of melted soft agar/host poured over the surface. Phage is then allowed to run over hardened surface. Phage may also be added directly to melted soft agar before pouring as described above.
- After 24 hours incubation, or when lysis is observed, the soft agar is scraped off the surface of the agar plates. Centrifuge at about 1000 rpm for 25 minutes to sediment the cellular debris and agar. Conserve the supernatant.
- This supernatant is passed through a .22 µm Millipore filter and the filtrate stored at 4°C to 8°C. Lysates should remain viable under refrigeration for long periods. They may also be frozen with or without cryoprotectant. If available, liquid nitrogen storage is the best method for long term storage. Most phage can also be freeze dried. ATCC uses double strength skim milk mixed half-and-half with the filtrate. (See: NOTE 4)
- Bacteroidaceae, Anaerobic
Many Bacteroidaceae species require anaerobic conditions for growth. ATCC recommends using pre-reduced media that was either freshly prepared or previously prepared and stored under anaerobic conditions.
Media can be prepared with reducing agents and stored in anaerobic environments such as anaerobic chambers. To prepare pre-reduced media, add 0.1 mL of a reducing agent for each 5-10 mL of mixture and let it sit for a minimum of 30 minutes. Any of the following reducing agents are appropriate: 1.5% Na2S*9H2O, 3% cysteine, or 5% coenzyme M.
Pre-reduced Anaerobically Sterilized (PRAS) media is also available commercially. Under anaerobic conditions, the media is boiled free of molecular oxygen, a reducing agent is added, and the media is then autoclaved and dispensed.- Rehydrate the vial contents under anaerobic conditions with 0.5 mL of the recommended broth medium.
- Aseptically transfer this aliquot to 5 mL of the same broth. At this time, you may inoculate an agar slant of the same medium and a blood agar plate, using 0.1 mL of the cell suspension for each.
- Incubate the broth and slants at 37°C under anaerobic conditions.
- Anaerobic conditions may be obtained by the methods described in the previous chapter, “Bacterial Growth and Propagation.”
- Incubate the blood agar plates anaerobically for colony formation or aerobically for aerobic contamination check.
- After an appropriate incubation period, which is strain dependent, growth should be evident by turbidity in the broth and by colonies on the anaerobic agar surfaces. An aerobic blood plate should show no growth.
- Bdellovibrio spp.
Members of the Bdellovibrio genus (eg, ATCC 27047) are host-specific predators of Gram-negative bacteria. Therefore, it is necessary to establish a growing culture of the host strain before opening the vial of the predator. Below, we describe a basic procedure on how to culture Bdellovibrio. - In advance, culture the appropriate host strain according to the recommended growth conditions.
- Rehydrate the freeze-dried pellet of Bdellovibrio spp. with 0.5 mL of the recommended medium and transfer the suspension to a sterile test tube.
- Add approximately 0.5 mL of the growing host strain.
- Melt the agar in tubes containing 2.5 mL each 0.6% semi-solid agar and place the tubes in a water bath set at 45°C.
- Add 0.2-0.3 mL of the host/predator suspension to each tube of melted agar and pour over the surface of a pre-warmed plate of recommended agar medium.
- Incubate plates aerobically at 30°C for 2-3 days. Observe daily until plaques or clearings are seen.
- Scrape off the overlay into a test tube and centrifuge at low speed to sediment the agar and cellular debris. The Bdellovibrio should be in the supernatant. Check for tiny, rapidly moving cells under a wet mount.
- Borrelia burgdorferi
B. burgdorferi is a fragile, sensitive organism that must have the appropriate medium and conditions for growth. The supplementation of rabbit serum to the required Barbour-Stoenner-Kelly (BSK) medium is essential for the growth of this organism. Additionally, fresh medium enhances growth, and medium older than one month should not be used. - Thaw and aseptically transfer the entire contents of the liquid nitrogen vial to a tube containing 5-6 mL of fresh recommended medium. Mix well.
- Transfer one-tenth of the cell suspension to two or three other tubes of fresh medium.
- Incubate the bacteria at 32°C to 37°C under microaerophilic conditions.
- Growth usually occurs after 48 hours, although some strains may take several days to grow. Acid formation during growth will change the medium to a light or yellowish orange color. Turbidity is not evident. Cells can be monitored under dark-field microscopy as long spiral rods with twitching motility.
- Leptospira spp.
Leptospira is an aerobic strain that is fairly difficult to culture. ATCC recommends culturing this bacterium in Leptospira Medium supplemented with agar (ATCC medium formulation 1470). (See: NOTE 5) - Aseptically withdraw the cell suspension from the thawed vial and inoculate a 10 or 12 mL tube of recommended medium, inserting the pipette just below the surface of the semi-solid medium. Aliquots of 0.5 mL from this tube may be used to inoculate additional tubes if needed. An aerobic blood plate may also be streaked to test for purity.
- Incubate test tubes with screw-caps slightly tightened with the blood plate aerobically at 30°C.
- Initial growth may take from 6 to 20 days depending on the strain. Observe growth as a slight tight band forming just below the surface of the medium. The band thickens as incubation continues. Cellular morphology under dark-field microscopy shows active spiral cells with flexing motility. The aerobic blood plate should show no signs of growth.
- Campylobacter spp.
Campylobacter species are often difficult to grow and maintain. Many Campylobacter species, such as C. concisus, C. mucosalis, and C. showae, require a Brucella albimi broth or trypticase soy agar medium which has been supplemented with formate and fumarate. - Rehydrate the contents of the vial with 0.5 mL of the recommended medium. Aseptically transfer the suspension to 5.0 mL of the same broth.
- Inoculate prepared agar plates, agar slants, or broth tubes with 0.1 mL aliquots of the suspension. Screw caps on tubes loosely to allow for gas exchange.
- Incubate the cultures at 37°C under microaerophilic conditions using an anaerobe jar and a Campylobacter microaerophilic gas generator with an active catalyst or any other suitable system which gives a final atmospheric mixture of 3-5% oxygen and 10% carbon dioxide.
- Initially, 3-7 days of incubation may be required before visible growth is evident. Later subcultures should require only 2-3 days.
- Helicobacter spp.
Helicobacter species have been isolated from a variety of animals, including humans. They require microaerophilic growth conditions and can be grown on trypticase soy agar supplemented with 5% defibrinated sheep blood. Below, we describe the biphasic method of Helicobacter revival. This method provides the best recovery and most rapid growth as compared to other methods of propagation. - Add 0.5 mL of the recommended broth medium to the vial.
- Mix the contents with the tip of the pipette until the pellet is rehydrated.
- Add 0.4 mL of the suspension to a fresh agar slant of recommended medium. Add the remaining 0.1 mL to a fresh agar plate.
- Place the plates and test tube slant cultures into an anaerobe jar with an active catalyst and a microaerophilic gas generator Gas Pak or within an incubator that can be set up for microaerophilic conditions (3-5% oxygen and 10% carbon dioxide).
- Incubate cultures at 37°C for 72 hours.
- After 72 hours of incubation, you should observe small colonies on the surface of the agar plate and slant. There should be heavy growth in the liquid portion of the biphasic slant.
- Legionella pneumophila
L. pneumophila strains are nutritionally fastidious and very sensitive to the caliber of the recommended growth medium, charcoal-yeast extract (CYE) Medium (buffered). ATCC recommends that the pH of the medium be checked when the temperature of the medium is cool. Additionally, the medium should be stored in the dark as exposure to light may result in the accumulation of peroxides, which may be inhibitory to bacterial growth. - Store vials at 4°C until ready to use.
- Rehydrate the freeze-dried material with 0.5 mL of the recommended broth.
- After the pellet has dissolved, transfer the vial contents to a test tube containing 5-6 mL of broth. Aliquots of this broth may be transferred to additional tubes of broth, to agar slants, or onto agar plates for purity and viability checks.
- Incubate the cultures at 37°C under atmospheric conditions of 5% carbon dioxide. Strains should show growth after 48 hours.
- Mollicutes
Mollicutes are a class of bacteria that are distinguished by their lack of a cell wall. Rather, most members of this class contain sterols in their cell membrane to increase membrane rigidity. Some genera included in this category include Mycoplasma, Spiroplasma, and Ureaplasma. Bacteria in this class tend to require a rich growth medium and are often sensitive to overgrowth. - Using the recommended medium, pipette 2.0 mL of broth into a sterile tube. Prepare additional tubes, each containing 4.5 mL of the broth.
- Open the freeze-dried vial as directed. Dissolve the pellet using approximately 0.5 mL of the broth from the tube containing 2.0 mL to the freeze-dried pellet. Withdraw the suspension and transfer it back into the tube.
- Perform a serial dilution by transferring 0.5 mL from the first tube to a tube with 4.5 mL, and then 0.5 mL from the second tube to a third tube, etc. This dilution series is important for titering purposes as well as to keep the culture in varying stages of growth. Many mollicutes strains die out when cultures become alkaline.
- Incubate the tubes under the recommended culture conditions and temperature for each strain. The incubation period will differ between strains. For strains requiring anaerobic conditions, ATCC recommends that an anaerobe jar or other appropriate procedure is used.
- Green and Purple Sulfur Bacteria
Green and purple sulfur bacteria are anaerobic or microaerophilic groups of bacterial strains that are capable of photosynthesis. Unlike algae, they do not use water as their reducing agent; rather, they use hydrogen sulfide. - Transfer the growing culture to fresh recommended medium, filling each 125 mL screw-capped bottle to the top.
- Incubate the culture at room temperature under incandescent light of 2,000-3,000 lux until the elemental sulfur (i.e. milkiness) that formed disappears.
- Feed the cultures with 1.0-2.0 mL of sterile neutral sulfide solution.
NOTE 3
Spotting the phage on plates makes visualizing the lysis easier. If phage is added directly to soft agar before pouring plates, hazy or tiny plaques may be difficult to see. Resistant host bacteria may also mask plaque formation.
To propagate phage:
NOTE 4
Broth propagation methods may also be employed with most phage. Unless otherwise noted in the product sheet, ATCC uses the Adams agar overlay method as described in M. H. Adams’ Bacteriophages2 for routine phage production.
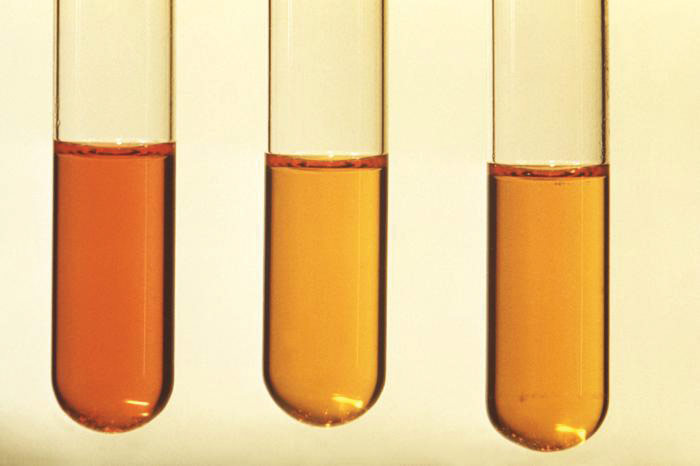
NOTE 5
ATCC provides the formulations for media for each product. Please note that most microbial media are not currently sold at ATCC.
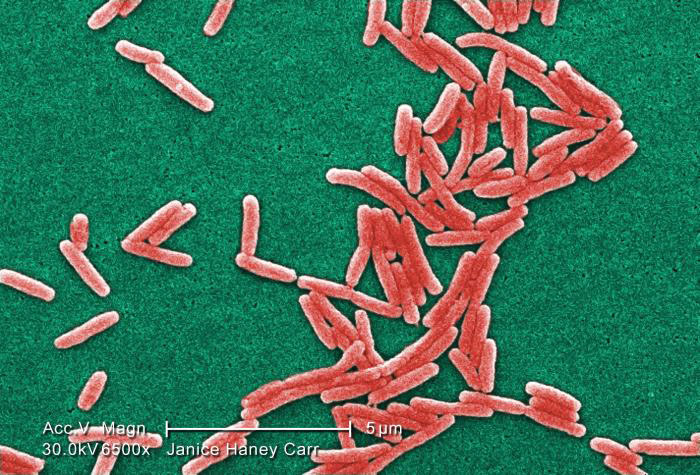 Image of Gram-negative Legionella pneumophila bacteria courtesy of Janice Haney Carr, CDC
Image of Gram-negative Legionella pneumophila bacteria courtesy of Janice Haney Carr, CDCBacterial Titering
Bacterial cell counts are necessary in order to establish or monitor bacterial growth rates as well as to set up new cultures with known cell counts.4 Bacterial cultures can be titered via determining the number of colony forming units per milliliter (CFU/mL) or by measuring the optical density at a wavelength of 600 nm (OD600).
To enumerate CFU/mL within a bacterial suspension at a given time-point, remove an aliquot from an actively growing solution and perform a serial dilution in an appropriate broth medium (See: NOTE 6). The extent of the dilution will depend on the growth rate and phase of the strain. Each dilution should be spread-plated onto several plates consisting of an appropriate agar-based medium, then grown under optimal conditions. Following a suitable growth period, count the number of colonies that have grown on each plate. For best results, only use plates harboring colony counts between a range of 25-250 colonies as this will provide an accurate representation of the bacterial titer. To determine the CFU/mL, calculate the average of the number of colonies from one dilution series then divide by the final dilution on the plate. For example, if you have three counts of 40, 37, and 43 from a set of plates at final of dilution of 10-7, the bacterial titer would be 4.0 x 108 CFU/mL.
NOTE 6
To ensure that the bacterial concentration of the original culture remains constant while determining titer, growth can be suspended at 4°C. Note that at 4°C, culture viability may be compromised, or alternatively, some bacterial strains may continue to grow slowly.
A spectrophotometer is also used to titer bacterial suspensions though determining the optical density, or absorbance, of a sample. The concentration of a bacterial culture is measured by projecting a beam of light, at a single wavelength of 600 nm, through the suspension within a transparent cuvette. When the beam of light passes through the sample, some light is absorbed while remaining light is quantified by a photometer. Generally, the more concentrated a solution is, the more light will be absorbed, and the higher the OD measurement will be. To ensure that the OD600 reading generated by the spectrophotometer is the OD of the bacterial suspension and not that of the medium, blank the machine before use with a medium-only control.
The growth rate and culturing requirements of bacteria can vary drastically between species, thus making it difficult to quantify a bacterial titer. For more information on how to culture a particular strain, see the product detail page. Additional information can also be found in Bergey’s Manual of Systematic Bacteriology 2nd Edition.1 (See: NOTE 7)
NOTE 7
Several bacterial genera, such as Borrelia and Leptospira, are not normally cultured on agar-based media. For these strains, ATCC uses light microscopy to establish titer.

Growth Media
Bacterial Culture Media
Depending on their composition or use, culture media can be categorized into several groups; these include defined, complex, selective, and enrichment medium. In a defined medium, the exact chemical composition is known. These types of media are usually composed of pure biochemicals, and are often used to study the minimal nutrient requirement of a microorganism. In contrast, the exact chemical composition of a complex medium is not known. This latter medium type often contains reagents of a biological origin, such as yeast extract and peptone, where the exact chemical composition is unknown. Complex media usually provide a large range of growth factors that assist in the cultivation of unknown and fastidious bacterial species.3
Media may also be formulated as selective or enriched. A selective medium is formulated to inhibit the growth of certain bacterial species and/or promote the growth of a specific species. These media can consist of additional selective reagents, such as high salt concentration to select for halophiles, or can be used under selective growth conditions. An enrichment medium also allows for the growth of specific bacterial species; however, enrichment media are supplemented with a reagent that permits, rather than inhibits, the growth of a particular species.3
Generally, bacterial culture media are mixtures of proteins, salts, trace elements, amino acids, and carbohydrates. The presence and volume of these components can vary significantly among bacterial species depending on the macro- and micro-nutrient requirements of each strain. The manner in which bacterial strains are cultured also varies widely. Liquid media are often used for the growth and propagation of pure batch cultures, while solid agar-based media are used for the isolation of pure cultures.
In addition to supplying nutrients, liquid medium can assist in the maintenance of pH. The pH can be sustained through one or more buffering systems such as 3-(N-morpholino)propanesulfonic acid (MOPS) or potassium phosphate. The osmotic environment can be maintained through the addition of salts, such as sodium chloride. ATCC uses numerous types of media in order to provide the optimal growth conditions for each bacterial species. The formulations for these media can be found on the ATCC website. (See: NOTE 5). Several bacterial media commonly used by ATCC include the following:
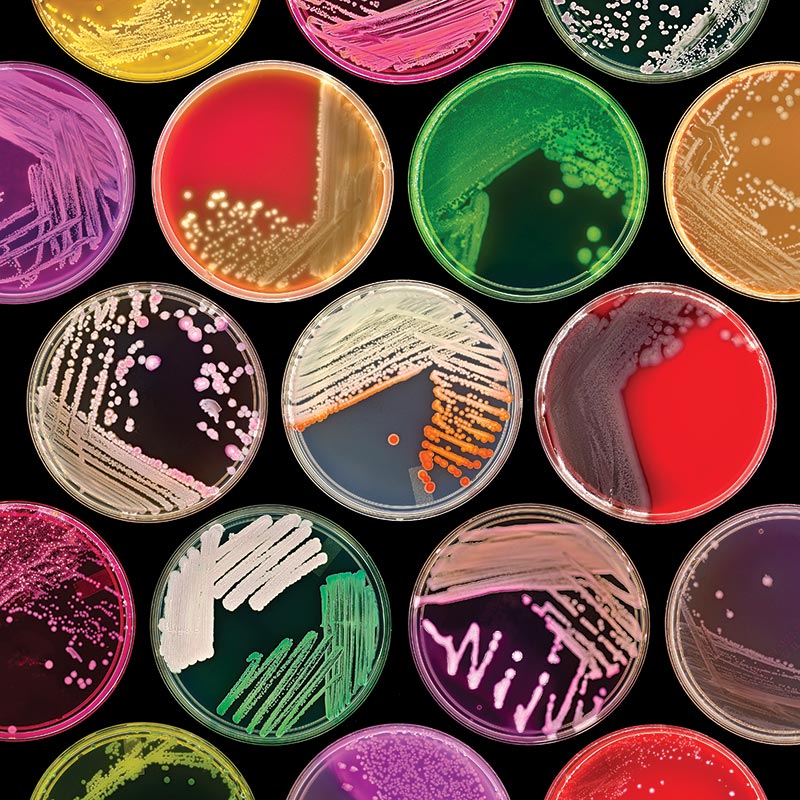 Image courtesy of Norm Baker, MS, MA, RBP, Johns Hopkins Univ. School of Medicine
Image courtesy of Norm Baker, MS, MA, RBP, Johns Hopkins Univ. School of MedicineBrain Heart Infusion Agarose (BHI) (ATCC medium formulation 44) is a complex, nutrient-rich, general-purpose growth medium used for culturing fastidious and nonfastidious microorganisms, including streptococci and pneumococci. It is generated from the dehydrated infusions of bovine or porcine brain and heart tissue. This medium can be supplemented with sodium chloride and disodium phosphate for osmotic and pH maintenance, respectively. ATCC currently uses both BHI agar and broth medium supplied from Becton, Dickinson and Company (BD).
Gonococcal (GC) Medium (ATCC medium formulation 814) is a growth medium used for the cultivation of Neisseria gonorrhoeae and other fastidious organisms. It can be employed as a basal medium in the preparation of Chocolate Agar, Thayer-Martin Medium, Martin-Lewis Agar, and Transgrow Agar. GC medium is prepared from a mixture of GC agar base, Dried Bovine Hemoglobin, and IsoVitaleX available from BD. For the selective growth of specific organisms, this medium can be supplemented with antibiotics including chloramphenicol, streptomycin, tetracycline, and ampicillin.
Haemophilus Test Medium (HTM) (ATCC medium formulation 5129) consists of a complex mixture of yeast extract, Mueller Hinton Broth, Porcine Hematin, and nicotinamide adenine dinucleotide (NAD). The main component, Mueller Hinton Agar, was originally prepared as a solid media used to test the antimicrobial susceptibility testing of common, non-fastidious, rapidly growing organisms. This initial media, however, was not suitable for fastidious organisms such as streptococci, gonococci, and Haemophilus species.5 HTM was subsequently developed for the testing of fastidious organisms.
Luria-Bertani Broth Medium (LB) (ATCC medium formulation 1065) is an all-purpose medium used by ATCC for the propagation and maintenance of Escherichia coli for molecular biology applications. It is prepared in the Miller Composition using an LB broth mixture supplied by BD, which consists of tryptone, yeast extract, and sodium chloride. LB can also be prepared as a solid medium through the addition of agar. This medium is often supplemented with various antibiotics for the maintenance of plasmid DNA.
Marine Broth (ATCC medium formulation 2) is a selective, complex growth medium used for the cultivation of heterotrophic marine bacteria. The marine environment offers a unique set of growth conditions consisting of high salinity and low temperatures. To simulate this environment, marine broth is formulated with peptone, yeast extract, and a high salt content. ATCC generates this medium using the Marine Broth 2216 preparation from BD. Marine broth may also be prepared as a solid medium through the addition of agar.
Middlebrook 7H9 Broth (ATCC medium formulation 2714) is formulated to support the growth and propagation of mycobacterium species. This medium is prepared as a mixture of Middlebrook 7H9 broth, glycerol, sodium pyruvate, and albumin-dextrose-catalase (ADC) enrichment.
Middlebrook 7H10 Agar (ATCC medium formulation 173) is formulated for the isolation and cultivation of mycobacteria from clinical and non-clinical specimens. This medium is prepared as a mixture of Middlebrook 7H10 broth, glycerol, and oleic acid-albumin-dextrose-catalase (OADC) enrichment. It differs from Middlebrook 7H9 broth in concentration of several salts as well as the addition of OADC enrichment rather than ADC enrichment. This medium is based on an improved formulation that was developed to promote the early growth of mycobacteria in vitro.6
Modified Chopped Meat Medium (ATCC medium formulation 1490) is a non-selective, complex medium that supports the growth of most spore-forming and non-spore-forming obligate anaerobes. This medium consists of a complex mixture of ground beef (fat-free), sodium hydroxide, trypticase peptone, yeast extract, Hemin, dipotassium phosphate, and vitamin K1 solution. Depending on the required growth conditions of each anaerobic strain, ATCC supplements this medium with various reagents such as glucose, arginine, or a combination of formate and fumarate.
Nutrient Agar/Broth (ATCC medium formulation 3) is a general-purpose medium for the cultivation of non-fastidious bacterial strains. ATCC prepares this media from a dehydrated stock provided by BD, which consists of beef extract and peptone. This medium can be further enriched by the addition of heart infusion broth. Alternatively, this medium can also be used for the selective growth of specific strains through the addition of varying concentrations of sodium chloride.
Reinforced Clostridial Medium (ATCC media formulations 1053, 2107) is used by ATCC for the cultivation and recovery of anaerobes, particularly Clostridium species, from a variety of sources. ATCC prepares this medium using dehydrated culture media from either Oxoid Limited or BD. This media consists of a complex mixture of salts, tryptose, beef extract, yeast extract, dextrose, starch, and L-Cysteine HCl.
Tryptic Soy Agar (ATCC medium formulation 18) is a medium used for the isolation and propagation of a variety of bacterial strains. ATCC often prepares this medium using a tryptic soy base, provided by BD, supplemented with 5% defibrinated sheep’s blood (ATCC medium formulation 260). The tryptic soy base mixture consists of a combination of tryptone, soytone, and sodium chloride. The addition of sheep’s blood helps to facilitate the growth of more fastidious bacteria.
Microbial Media Formulations
ATCC provides the formulations for media for each product. Please note that most microbial media are not currently sold at ATCC.
Media Ingredients
IsoVitaleX
IsoVitaleX (BD) is a chemically defined enrichment additive used for the cultivation of fastidious bacteria. This reagent is used in lieu of yeast concentrate to function as a nutrient supplement for the growth of bacteria including gonococci and Haemophilus. IsoVitaleX consists of a mixture of Vitamin B12, L-Glutamine, adenine, guanine hydrochloride p-Aminobenzoic acid, nicotinamide adenine dinucleotide, thiamine pyrophosphate, ferric nitrate, thiamine hydrochloride, L-cysteine hydrochloride, L-cysteine, and dextrose.
Nicotinamide adenine dinucleotide (NAD)
NAD is a dinucleotide compound found in all living cells. It functions as a coenzyme involved in oxidation/reduction reactions, transporting electrons from one reaction to another. NAD can also be used as a substrate in several biochemical reactions such as mono- and poly-ADP-ribosylation.7 The addition of NAD to bacterial culturing medium provides an essential role in metabolism as a coenzyme in redox reactions and can additionally function as a substrate for bacterial DNA ligases.8
Peptone
Peptone is a water-soluble protein derivative used in bacteriology culture media. This reagent is prepared via the partial hydrolysis of an animal protein by an enzyme or acid. Generally, not all bacterial species can use free atmospheric nitrogen. Many species require either organic or inorganic fixed nitrogen.9 Peptone is used in bacterial media as an organic source of nitrogen and is often used in serum-free medium. The nutritional value of peptone is dependent on the amino acid content that supplies the essential nitrogen. The starting material for peptone can range from animal to plant; these can include meat, soybean, casein, and whey.10
Yeast Extract
Yeast extract is prepared as a water-soluble extract of autolyzed Saccharomyces cerevisiae yeast cells. During autolysis, endogenous yeast digestive enzymes break down protein content into peptides and amino acids which can be used by bacteria as a source of nitrogen. Additionally, yeast extract provides an essential source of water-soluble B-complex vitamins, carbohydrates, and free glutamic acid.11, 12
ADC
ADC enrichment is used in Middlebrook 7H9 media for the selective growth of mycobacteria. It is formulated with sodium chloride, bovine albumin (fraction V), dextrose, and catalase. The addition of dextrose provides an extra source of energy. Both the albumin and catalase have protective roles via binding free fatty acids which may be toxic to mycobacteria and ridding the environment of toxic peroxides, respectively.12
OADC
OADC is an enrichment supplement added to Middlebrook 7H10 agar for the cultivation of mycobacteria. This additive provides the same reagents as ADC in addition to oleic acid. Addition of OADC provides faster and more robust growth of Mycobacterium species.6, 12
Sodium Pyruvate
Pyruvate is an intermediary organic acid metabolite produced as an end product of glycolysis and the Entner-Doudoroff Pathway.13 The addition of sodium pyruvate in bacterial culture media provides both an energy source and a carbon skeleton for anabolic processes. This reagent has also been shown to provide protection against hydrogen peroxide.14
Dextrose
Dextrose is a highly nutritious supplement used for the cultivation of fastidious organisms, older cultures, and cultures with small inoculums. This reagent can be used as a dextrose broth/agar medium or as a supplement within a complex or defined medium. Generally, dextrose provides bacteria with a carbon source, and can be supplemented with 5% blood to provide additional growth factors for fastidious micro-organisms.15
Media Supplements
The growth media recommended for some bacterial species may require the addition of components not already available in the base medium. These components may include antibiotics, serum, blood, or chemical supplements.
After supplements have been added to the base medium, the shelf life of the medium should be determined on a case-by-case basis. Media containing antibiotics tend to degrade faster than base media alone. Media containing supplements should not be frozen as this may cause certain compounds to precipitate out of solution; media should be stored at 2°C to 8°C. For additional information regarding the preparation, storage, or usage of specific additives, contact your local supplier or consult with the manufacturer’s product information sheet.
Chemical Supplements
- Trace Elements
Trace elements are micronutrients required by microorganisms in small amounts. These nutrients are metal ions required by most microorganisms for survival as they usually function as cofactors for essential enzymatic reactions. It is often not necessary to provide trace elements in media as they can be present in minute amounts within the water or other media reagents. Trace elements are, however, added to minimal media as it contains only the minimum addition of nutrients required for microbial growth. Examples of trace elements required for bacterial nutrition include zinc, copper, manganese, molybdenum, and cobalt.3 ATCC offers a Trace Mineral Supplement (ATCC MD-TMS) that is based on Wolfe’s mineral solution. - Salts
Various salts, such as magnesium, calcium, iron, and potassium salts, are required for bacterial growth as they provide major elements that can function as cofactors for certain enzymatic reactions. In addition to the aforementioned function, calcium and iron form major components of endospores and cytochromes, respectively.3 Furthermore, the presence of salts, particularly sodium chloride, within a medium assists in the maintenance of osmolality. Most bacteria require either an isotonic or hypotonic environment for optimum growth. In these environments, water either flows in and out of the cell at equal rates or water only flows into the cell, respectively.3 - Glucose
Microbial media are often supplemented with glucose as a source of carbohydrates. In addition to this nutritional role, glucose may also be added as a means to repress an array of inducible enzymes in many different bacterial species. This operon repression is accomplished via the inhibition of cyclic AMP (cAMP) synthesis. The activity of adenylate cyclase, the enzyme required for cAMP synthesis, is blocked in the presence of glucose. In bacterial species such as Escherichia coli, this inhibition requires the cells to use glucose as the primary energy source rather than metabolizing poorer sources of energy.3 - Blood
Blood is often added to growth medium to enhance the cultivation of highly fastidious microbial species. Examples of such media include Tryptic Soy Agar and Dextrose Agar, both of which are often supplemented with 5% defibrinated sheep blood. The enrichment of a medium with blood supplies nutrients, such as growth factors, as well as assists in the determination of hemolytic reactions and pigmentation.12

Antibiotics
Antibiotics can be added to bacterial culture media to select for the growth of a specific variant. This type of medium is often used for the selection of strains that have inherent antibiotic resistance, harbor plasmids, or are genetically engineered.3 The addition of antibiotics often decreases the shelf life of a medium as compared to the base medium. To extend the shelf life, media containing antibiotics should be stored at 2°C to 8°C in the dark.
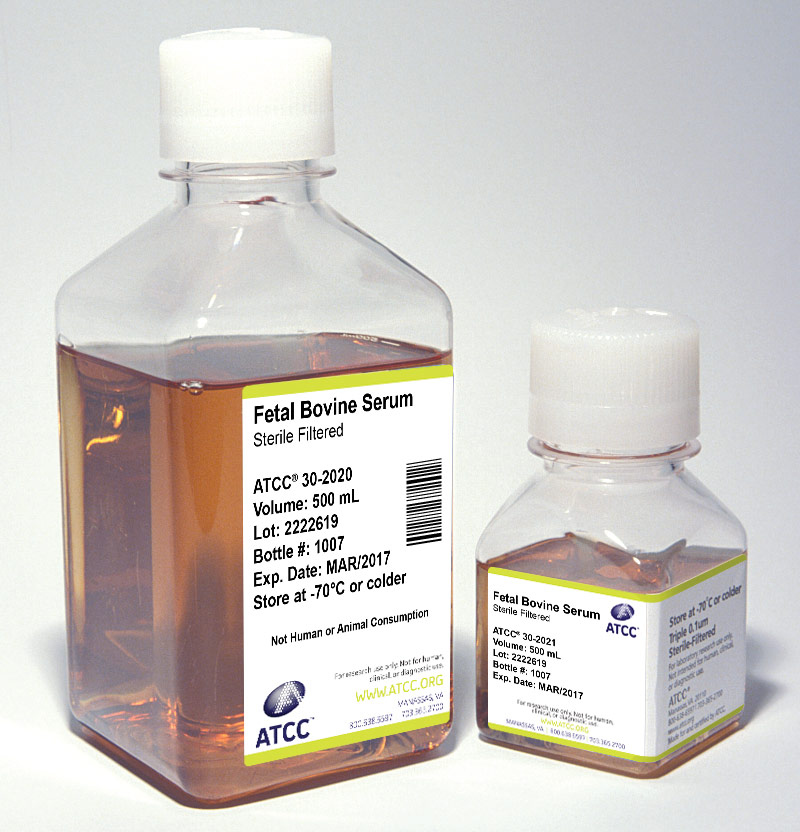
Serum
Sera can serve as a source of growth factors, proteins, vitamins, hormones, carbohydrates, lipids, amino acids, minerals, and trace elements. Additionally, serum can function as a pH buffer and can inactivate proteolytic enzymes. The exact composition of sera is unknown and varies from lot to lot, although lot-to-lot consistency has improved in recent years.
Sera from rabbit, horse, fetal bovine, and calf bovine sources are used to support the growth of some bacterial species in culture. Many pathogens, such as members of the genera Corynebacterium, Borrelia, Leptospira, and Mycoplasma, require the addition of serum to support growth. Media formulated to support these aforementioned strains are commonly supplemented with heat-inactivated rabbit or horse serum.
Unfortunately, naturally derived products from animals, such as sera, may contain adventitious microorganisms. All reputable suppliers routinely test their products for infectious virus by several methods including fluorescent antibody, cytopathic effect, and hemadsorption. These products are also screened for the standard microbial contaminants such as bacteria, fungi, and mycoplasma. To reduce the risk of any possible contamination, ATCC recommends that all serum should be triple filtered through 0.1 µm filters before use.
ATCC offers the following three types of animal sera:
- Fetal Bovine Serum (also known as fetal calf) – ATCC 30-2020
- Fetal Bovine Serum qualified for embryonic stem cells – ATCC SCRR-30-2020
- Iron-supplemented Calf Bovine Serum – ATCC 30-2030
These products are rigorously tested for adventitious infective agents and sourced only from U.S. herds. Furthermore, each lot is tested for its ability to support cell growth and is the same sera used in ATCC labs.
- Storage
Store sera at -20°C or colder for storage over 30 days. ATCC sera are routinely stored at -70°C. Do not store sera at temperatures above -20°C for any length of time. Avoid repeated freeze-thaws by dispensing and storing sera in aliquots. - Thawing
The following procedure is used to thaw serum: - Place the frozen serum in a refrigerator at 2°C to 8°C overnight.
- Put the bottles in a 37°C water bath and gently agitate occasionally to mix the solutes that tend to concentrate at the bottom of the bottle.
- Turbidity and precipitates
All sera may retain some fibrinogen. Because external factors may initiate the conversion of fibrinogen to fibrin, flocculent material or turbidity may be observed after the serum is thawed. The presence of this material does not alter the serum’s performance. If the presence of flocculent material or turbidity is a concern, it can be removed by filtration through a 0.45 µm filter.
A precipitate can form in serum when incubated at 37°C or higher for prolonged periods of time. This is often mistaken for microbial contamination. This precipitate may include crystals of calcium phosphate, but this does not alter the performance of the serum as a supplement. Heat inactivation of sera can also cause the formation of precipitates. - Heat Inactivation
Because serum is a blood component, it contains complements, which are a group of proteins that are part of the immune response. These proteins can lead to complement-mediated cell lysis of bacterial cells. To reduce the risk of bacterial lysis, serum used in microbial media can be heat inactivated. Heat inactivation, however, will reduce or destroy growth factors present in the serum; this may be detrimental to the growth of some microbial cells.
The following procedure can be used to heat-inactivate serum: - Thaw the serum.
- Preheat a water bath to 56°C. Use sufficient water to immerse the bottle above the level of serum.
- Mix the thawed serum by gentle inversion and place it in the 56°C water bath. The temperature of the water bath will drop.
- When the temperature of the water bath reaches 56°C again, continue to heat for an additional 30 minutes. Mix the serum gently every 5 minutes to ensure uniform heating.
- Remove the serum from the water bath, and cool quickly. Store the serum at -20°C or colder.

Preservation
Most bacterial strains can be stored for many years as freeze-dried cultures (lyophilization) or at temperatures below -130°C (cryopreservation).4 There are many advantages of preservation that far outweigh the required investment in the equipment and reagents needed to maintain living cultures. These advantages include:
- Overall safety of bacterial stocks against loss due to equipment failure or contamination by other microbial organisms.
- Elimination of time, energy, and material costs associated with the maintenance of bacterial strains not currently in use.
- Insurance against phenotypic drift associated with prolonged passage, due to genetic instability and/or selective pressures.
- Creating a standard working stock that can be used for a series of experiments.
Cryopreservation
Overview
Generally, freezing a suspension of living cells may result in a decrease in viability.16, 17 As the bacterial cell suspension is cooled below the freezing point, ice crystals begin to form and the concentration of solutes in the suspension increases. The formation of intracellular ice crystals can result in the damage of cellular structures. This effect can be minimized if water within the cells is allowed to escape by osmosis during the cooling process. A slow cooling rate, generally -1°C to -10°C per minute, facilitates this progression. However, as the cells lose water, they shrink in size and will quickly lose viability if they surpass a minimum threshold volume. The addition of cryoprotectant agents, such as glycerol or dimethylsulfoxide (DMSO), will mitigate these effects.18, 19 For the preservation of bacterial cultures, ATCC recommends using glycerol as a cryoprotectant.
The standard procedure for cryopreservation is to freeze cells slowly until they reach a temperature below -70°C in medium that contains a cryoprotectant. Vials are then transferred to a liquid-nitrogen freezer to maintain cultures at temperatures below -130°C.
The recovery of cryopreserved cells requires the rapid thawing of the bacterial suspension in a 37°C water bath. The entire contents of the vial are then transferred to an appropriate growth medium.
There are numerous factors that can affect the viability of recovered bacterial strains. These critical parameters can include the composition of the freeze material, the growth phase of the bacterial strain, or the concentration of bacterial cells within the solution. To obtain optimal cell viability upon recovery, modify the cryopreservation procedure for each bacterial strain, being sure to harvest cultures during the late logarithmic phase of growth.
Contact ATCC for more information on the cryopreservation of bacterial strains. ATCC bacterial strains that are frozen are commonly cryopreserved with a medium consisting of a final concentration of 10% sterile glycerol.
Freeze Medium
Glycerol and DMSO are the most common cryoprotectant agents. ATCC predominantly recommends using a 20% glycerol stock at a final concentration of 10%. However, if the bacterial strain is sensitive to glycerol, a 50% DMSO stock can be used at a final concentration of 5%. In contrast, glycerol can be sterilized via autoclavation whereas DMSO must be sterilized by filtration. Care should be used when handling any DMSO solution as it will rapidly penetrate intact skin and may carry toxic contaminants along with it.
Use only reagent-grade DMSO or glycerol. Store both in aliquots protected from light. ATCC offers DMSO (ATCC 4-X) that has been thoroughly tested for use, as well as TSB with 10% glycerol (ATCC 20-2200) for the cryopreservation of non-fastidious bacteria. Overall, the optimum formulations for individual bacterial strains must be determined empirically.
Equipment
- Cryopreservation vials
There are two materials to choose from for cryopreservation vials: glass or plastic. Glass vials are more difficult to work with; they need to be sterilized before use, they need to be sealed with a hot flame, and they can be difficult to open. However, they are preferred for long-term storage (many years) of valuable cultures and are considered fail-safe once properly sealed.
If cryopreservation in glass ampoules is not possible, plastic vials can be used. Plastic vials come in two varieties: those with an internal thread and silicone gasket, and those with an external thread. Vials with an internal-thread were the first commercially available, but have some disadvantages over the external-thread version. For example, while the silicone gasket provides an excellent seal, it needs to be tightened just right; the vial will leak if the seal is too tight or too loose. For the storage of cryopreserved stocks, ATCC uses plastic vials. - Controlled-rate freezing chambers
There are several means to achieve a cooling rate of -1°C per minute. The best method involves the use of a computer controlled, programmable electronic freezing unit (such as Thermo Scientific* CryoMed Freezers), which rigorously maintains this rate of cooling. This is the method used exclusively at ATCC. Such equipment is relatively expensive and absolutely necessary for only the most sensitive strains.
A less costly approach is to place the cryopreservation vials into an insulated chamber and cool for 24 hours in a mechanical freezer at -70°C or colder. There are several commercially available freezing chambers which achieve a cooling rate very close to the ideal -1°C per minute (CoolCell LX; ATCC ACS-6000). Alternatively, the vials can be placed into a polystyrene box, with 15 mm (3/4 inch) thick walls and 1L capacity that is packed with paper, cotton wool, or foam peanuts for insulation.
Liquid Nitrogen Freezing Storage
The ultra-low temperatures (below -130°C) required for long-term storage can be maintained by specialized electric freezers, or more commonly, by liquid nitrogen freezers. There are two basic types of liquid nitrogen storage systems: immersing vials in the liquid or holding vials in the vapor phase above the liquid. The liquid-phase system holds more nitrogen and thus requires less maintenance. However, there is always a chance that some liquid will enter improperly sealed vials, which may then explode when retrieved. For this reason, ATCC strongly recommends storage in vapor-phase.
Vapor-phase storage systems create a vertical temperature gradient within the liquid nitrogen container. The temperature at the bottom of the container will be -196°C, whereas the temperature at the top will vary depending upon the amount of liquid nitrogen at the bottom and the length of time the container is open. To ensure the safe storage of cells, maintain sufficient levels of liquid nitrogen in the container so that the temperature at the top is -130°C or colder. All storage systems should be equipped with temperature alarms.
Cryopreservation Procedure
The procedure below will work for most non-fastidious bacterial strains and should be modified as needed. Additional information on the preservation of fastidious bacterial strains can be found in the latter segment of this chapter. Freeze medium formulations for ATCC bacterial strains can be found on the ATCC website.
- In preparation for freezing, grow the bacterial strain under optimal conditions in an appropriate medium as to retain the salient features of the strain. Bacterial strains should be grown to late log phase.
- When freezing bacteria, add 5 to 10% glycerol or DMSO in culture medium. Glycerol is usually prepared in aqueous solution at double the desired final concentration for freezing. It is then mixed with an equal amount of cell suspension.
- Label the appropriate number of vials with the name of the bacterial strain and the date. Aliquot 1 to 1.8 mL of the bacterial suspension to each vial and seal. Seal plastic ampoules tightly with the screw cap. Seal glass ampoules with a gas-oxygen torch, pulling the neck of the ampoule as it is rotated in the flame.
- Allow the cells to equilibrate in the freeze medium at room temperature for a minimum of 15 minutes but no longer than 40. After 40 minutes, cell viability may decline if DMSO is used as the cryoprotectant.
- Place the vials into a pre-cooled (4°C), controlled-rate freeze chamber and place the chamber in a mechanical freezer at -70°C (or colder) for at least 24 hours. Alternately, use a pre-cooled (4°C) programmable freezer unit set to cool the vials at -1°C per minute until a temperature below -40°C is achieved and then set the temperature to abruptly drop to -130°C.
- Quickly transfer the vials to liquid nitrogen or a -130°C freezer.
- Record the location and details of the freeze.
- After 24 hours at -130°C, remove one vial, restore the bacterial strain in culture medium, and determine the viability and sterility.
Recovery of Cryopreserved Cells
The frozen bacterial solution needs to be warmed as rapidly as possible and then immediately transferred to an appropriate growth medium. Some bacterial strains may take more time than normal to fully recover from cryopreservation.
- Prepare a culture vessel that contains at least 10 mL of the appropriate growth medium equilibrated for both temperature and pH.
- Remove the vial from the liquid nitrogen freezer and thaw by gentile agitation in a 37°C water bath (or a bath set at the normal growth temperature for that bacterial strain). Thaw the strain rapidly until all ice crystals have melted (approximately 2 minutes).
- Remove the vial from the water bath and decontaminate it by dipping in or spraying with 70% ethanol. Follow strict aseptic conditions in a laminar flow hood for all further manipulations.
- Unscrew the top of the vial and transfer the entire contents to the prepared growth medium.
- Examine the cultures after an appropriate length of time.
Lyophilization
Overview
Freeze-drying is a process where water and other solvents are removed from a frozen product via sublimation.20 Sublimation occurs when a frozen liquid goes directly to a gaseous state without entering a liquid phase. The freeze-drying process results in a stable, readily rehydrated product. This process consists of three steps: pre-freezing the product to form a frozen structure, primary drying to remove most water, and secondary drying to remove bound water.
During the initial freezing process, ice crystals begin to form and the concentration of solutes in the suspension increases. The method used during freezing can greatly affect the ability to freeze-dry the material. It is recommend using slow rates of cooling as this will result in the formation of vertical ice crystal structures, thus allowing for more efficient water sublimation from the frozen product.
Freeze-dried products are hygroscopic and must be protected from moisture during storage. Additionally, these products are sensitive to other factors including oxygen and temperature, which can significantly decrease the shelf life. It is important to store freeze-dried material in a manner that protects the product from exposure to moisture and oxygen, and to store the material at refrigerated temperatures (4°C).
Contact ATCC for more information on the lyophilization of bacterial strains. Most ATCC bacterial strains for distribution are prepared as freeze-dried cultures.
Equipment
- Lyophilization Vials
For the storage of freeze-dried organisms, ATCC uses both double-vialed glass ampoules and stoppered serum vials. Double-vialed glass ampoules are used for the lyophilization of batch cultures via a component freeze-dryer. Samples prepared within these vials are initially freeze-dried in cotton-plugged inner vials, which are then sealed in outer vials under vacuum. Outer vials are supplied with a small amount of a silica gel desiccant, which sustains a state of dryness. Samples preserved in this fashion can be stored indefinitely.
In contrast, stoppered serum vials are used for lyophilization using the Preceptrol method. This particular technique was developed as a way to produce a large number of vials in a cost-effective manner. Bacterial strains preserved in this manner are popular strains often used in quality control, teaching, and testing. In this method, material is freeze-dried in a glass serum vial then sealed with a rubber stopper and metal cap. Because these samples are usually distributed in serum vials capped with butyl rubber stoppers rather than the double-vialed ampoules, they are cheaper to produce and allow ATCC to reduce the usual fees. Samples prepared with the Preceptrol method have a shelf-life of approximately 5 years. - Lyophilization Apparatuses
There are several methods available for the lyophilization of bacteria. Two such methods employed by ATCC include the Preceptrol method and the use of a Component Freeze-dryer. In both freeze-drying procedures, samples are mixed with a suitable preservative, dispensed into the appropriate ampoule, and allowed to slowly freeze into a solid mass. Once frozen, samples are lyophilized within a freeze-drying system (VirTis Genesis Pilot Lyophilizer, Millrock Technology Max85 Freeze Dryer).
During the primary drying phase, water is removed from the frozen product via sublimation. This is accomplished through the use of a vacuum pump, which allows water molecules to migrate from the frozen product and condense on a moisture trap called a condenser. For this to be possible the temperature of the condenser must be colder than the product temperature; the difference in these temperatures will affect the rate of sublimation. When primary drying is complete, all residual moisture is removed by directly heating the product. During this secondary drying phase, water must be desorbed to a residual moisture content of 1% or less. This process requires a low pressure, low condenser temperature system. Once dried, the ampoules are properly sealed and stored at refrigerated temperatures (4°C).
Storage and Viability of Lyophilized Strains
To maximize the recovery of viable cells, bacterial cultures must be in optimum condition before the lyophilization process.21 Generally, the age of a culture can affect its ability to survive freeze-drying. Bacterial cultures that are harvested from late log phase or early stationary phase have the greatest chance for survival during low-temperature stresses. In addition to proper preparation of bacterial cultures, the recovery of viable cells may also be affected by the use of appropriate growth conditions and media during reconstitution.22 Nutritive, non-selective growth media best aids in maximizing recovery.23
Because lyophilized products are hygroscopic, they must be stored under moisture-free conditions. Exogenous factors such as oxygen content and storage temperature can affect the shelf-life of freeze-dried strains. Oxygen can negatively react with the product, affecting culture viability, and is directly proportional to storage temperature. Therefore, long-term storage of lyophilized products should be kept at refrigerated temperatures (4°C) under conditions that protect the sample from exposure to moisture or oxygen.

Lyophilization Procedure
- Component Freeze-dryer
- Wash inner vials (Glass Vials, Inc., 11.5 x 35 mm) and stopper with an oil-free cotton plug. Subsequently, label each vial and autoclave.
- Prepare outer vials (Glass Vials, Inc., 14.25 x 85.0 mm) by placing a small amount of silica gel granules (Fisher Scientific, grade 42, 6-16 mesh) in the vials to cover about half the bottom. Add a small cotton wad to cushion the inner vial. Heat this overnight at 100°C.
- Prepare the bacterial strain under optimal conditions in an appropriate medium as to retain the salient features of the strain. Bacterial strains should be grown to late log phase.
- Suspend cells in Reagent 20 (ATCC medium formulation 9520) and mix thoroughly. Dispense 0.2 mL of this suspension into each inner vial. Replace the cotton plug and trim it so that it is flush with the edge of the vial.
- Next, place the inner vials in a stainless steel pan and slowly freeze the sample.
- Once frozen, lyophilize the samples for 18 hours within a freeze-dryer. Before use, the condenser should be pre-chilled and the system evacuated to below 30 µm of Hg.
- When the lyophilization cycle is complete, remove the inner vials from the freeze-dryer and insert them into the prepared outer vials. Tamp a ¼ inch plug of fiber paper above the cotton-plugged inner vial. This should be performed in a dry cabinet.
- Subsequently, heat the outer vial using a torch, rotating the vial until the glass just above the fiber paper begins to constrict. This should create a narrow capillary tube. Once the glass cools, attach the vial to a port of a manifold. Evacuate the system to less than 50 µm of Hg.
- Seal the vials at the site of the capillary using a torch. Store vials at 4°C. After 24 hours, restore the bacterial strain in culture medium, and determine the viability and sterility.
- Preceptrol Method
- Label 2 mL serum vials (Wheaton Scientific). Place these vials into a tray, cover with a 6 ½ inch diameter filter and aluminum foil, and bake at 180°C for four hours. Meanwhile, autoclave the slotted butyl rubber stoppers (West).
- Prepare the bacterial strain under optimal conditions in an appropriate medium as to retain the salient features of the strain. Bacterial strains should be grown to late log phase.
- Suspend the cells in Reagent 18:
- 0.75 g Trypticase Soy Broth
- 10.0 g Sucrose
- 5.0 g Bovine Serum Albumin Fraction V
- 100 mL Distilled water
- Filter-sterilize through a 0.2 µm filter.
- Dispense 0.4 mL of the suspension into each vial and gently place a sterile stopper on each vial. Do not fully push stoppers in as vapor must be allowed to escape.
- Next, slowly freeze the sample in a mechanical freezer.
- Once the samples are frozen, cover the vials with sterile cotton, ½ to 1 inch thick. Then, lyophilize the samples for 18 hours within a freeze-drying system. Before use, the condenser should be pre-chilled and the system evacuated to below 100 µm of Hg.
- When the lyophilization cycle is complete, remove the vials from the freeze-dryer and spray thoroughly with a disinfectant such as Amphyl (Sterling Drug, Inc.). Place vials under ultraviolet light in a biological safety cabinet for at least 30 minutes.
- Seal the vials with aluminum caps (Wheaton) and store at 4°C. After 24 hours, restore the bacterial strain in culture medium, and determine the viability and sterility.
Recovery of Lyophilized Cells
- To rehydrate freeze-dried strains, add 0.3 to 0.4 mL of broth.
- Mix well and transfer to a tube containing 5 to 6 mL of broth medium.
- A few drops of this suspension may also be added to an agar slant or place. Growth medium should be chosen to maximize the recovery of cells. The optimum medium for each strain can be found on the product detail page for that specific material on the ATCC website.
- Incubate strains under the appropriate temperature and atmospheric conditions.
Preservation of Fastidious Bacteria
Most bacterial strains can be freeze-dried, and almost all strains can be cryopreserved and maintained in liquid nitrogen vapor. In preparation for cryopreservation or lyophilization, bacterial strains should be grown under optimal conditions. This can include growth on agar, in shaken broth cultures, or in static broth cultures. Additional care must be taken when preparing fastidious bacterial species for preservation.
Anaerobes
To properly preserve anaerobic bacteria, anaerobic conditions must be maintained during growth, harvesting, dispensing, and freezing. The cryoprotectant and suspending medium must be pre-reduced, and anaerobic conditions should be maintained with oxygen-free gas flow using a sterile cannula.
Bacteriophages
Most bacteriophages can be freeze-dried successfully; exceptions are stored in liquid nitrogen.24, 25 Prior to preservation, bacteriophages can be propagated in a soft agar layer or in broth; a titer of 108 pfu/mL is desirable. Bacteriophages can be cryopreserved in the absence of a cryoprotectant, performed under a controlled freezing rate. However, bacteriophages that require a cryoprotectant can be mixed with an equal volume of 20% glycerol. To freeze-dry, filtered bacteriophage suspensions should be mixed with 20% skim milk and lyophilized according to the component freeze-dryer method.
Mollicutes
Prior to cryopreservation or lyophilization, mollicutes should be grown in broth. Following growth, cell suspensions should be centrifuged and resuspended in equal amounts of broth and 20% glycerol. Alternatively, cells can be resuspended in equal amounts of broth and 24% sucrose for lyophilization in the component freeze-dryer method.26
Neisseria, Haemophilus, Campylobacter, and Helicobacter
These genera of bacteria are very sensitive to damage during preservation.27 To maximize cell viability and recovery, cells should be grown under optimal conditions and harvested at the proper point in the growth curve. To stabilize these strains during lyophilization, supplement the suspending medium with 0.5% sodium ascorbate prior to freezing.
Spirochetes
Spirochetes are very difficult to lyophilize. It is recommended that spirochetes be cryopreserved with 10% glycerol and stored in liquid nitrogen vapor.28
Special Hazards
Care must be taken during the cryopreservation or lyophilization of bacterial strains. Problems such as contamination, breakage of glass ampoules during handling and storage, dispersal of freeze-dried bacteria when opening glass ampoules, and the handling of liquid nitrogen must all be considered. To prevent contamination and the dispersal of bacteria, aseptic technique must be followed. This can include the decontamination of all equipment and vials as well as performing all preparations in a biological safety cabinet. Additionally, protective clothing should be worn during preparation to prevent contamination as well as to guard against harm due to contact with liquid nitrogen.

Biosafety and Disposal
Biosafety
The need for precautions when experimenting with bacterial cultures depends upon the source and nature of the biological material, the experimental procedure, and the laboratory/containment conditions. Since every situation is different, the risks need to be identified for each individual strain and the appropriate precautions need to be taken before any work begins.
More information on risk assessment and precautions can be found in the Center for Disease Control (CDC) publication Biosafety in Microbiological and Biomedical Laboratories.29 The text of this publication is available in its entirety on the CDC website at www.cdc.gov.
ATCC assigns a biosafety level (BSL) to each bacterial strain for the purposes of packaging for safe shipment. ATCC follows federal biosafety guidelines and takes several factors into consideration when assessing a potential hazard, and in some cases the ATCC assigned biosafety level is more restrictive. Generally, ATCC only ships and stores bacterial strains with a biosafety level assignment of 1, 2, or 3.
Biosafety Level 1
- Work involving well-characterized bacterial strains not known to consistently cause disease in immunocompetent adult humans.
- Work can be conducted on the bench top using aseptic technique, no special containment equipment or facility is required.
Biosafety Level 2
- Work involving bacterial strains that pose a moderate hazard to healthy adult humans.
- Work should be conducted in designated biological safety cabinets within laboratories with restricted access.
Biosafety Level 3
- Work involving indigenous or exotic agents that may cause serious or potentially lethal disease via inhalation.
- Work should be conducted in biological safety cabinets localized inside a specialized BSL-3 containment facility within laboratories with restricted access.
As the recipient of a bacterial strain, take into account not only the nature of the material but also the manipulations employed during its handling when assessing the potential laboratory risk. Keep in mind that there will be situations where the intended use of an agent may require more stringent precautions than associated with the assigned biosafety level.29
Disposal of Infectious Materials
All bacterial cultures, stocks, and potentially infectious materials need to be properly decontaminated prior to disposal. The written method for proper decontamination should be available in the laboratory and BSL facility. Several methods of sterilization include autoclavation, chemical disinfection, incineration, or any other validated decontamination method. More information on the disposal of bacterial cultures can be found in the Center for Disease Control (CDC) publication Biosafety in Microbiological and Biomedical Laboratories.29

Bacterial Authentication
When preparing a bacterial strain for distribution, ATCC performs numerous quality control (QC) assays to guarantee that the product is of the highest standard before it reaches the consumer. All bacterial strains undergo thorough phenotypic and genotypic examinations to ensure that strain identification is accurate, the culture is pure, and that all biochemical results are consistent. To assist in maintaining these standards, all equipment and biochemical tests required in the QC process are similarly evaluated for quality assurance. Described below are several tests that ATCC commonly performs on bacterial strains during QC analysis.
Phenotypic Characterization
When bacterial strains are initially received by ATCC, they are analyzed for characteristic colony and bacterial morphology; this is observed via colony growth and Gram staining, respectively. Additionally, samples are tested for culture purity on blood agar plates. The presence of more than one colony type is often, but not always, indicative of a mixed culture.
In addition to purity and morphology, strains are also examined for consistency in sugar metabolism, antibiotic susceptibility, and/or broad-spectrum biochemical reactivity. To confirm the bacterial species identity based on these associated phenotypic traits, ATCC commonly uses appropriate API strip tests and Vitek cards. Both aforementioned techniques employ over 20 different biochemical tests in reaction cells to assess the growth and viability of the bacterial strain.30 The API strip test characterizes bacterial identity through a manual, micro-method that generates a seven-digit code based upon the reaction of the individual biochemical assays. This generated code is the identifier number linked to a specific bacterial species. In contrast, the Vitek system provides an automated, computer-based method of species identification via the measurement of light attenuation associated with each biochemical reaction.30, 31 For this latter system, ATCC requires a ≥ 90% probability of organism identification.

Genotypic and Proteotypic Characterization
ATCC genotypically confirms bacterial species primarily through 16S ribosomal RNA (rRNA) sequencing. These sequences are commonly used to analyze bacterial phylogeny and taxonomy as they are highly conserved and universal among prokaryotic species. In general, the 16S rRNA gene sequence is composed of both variable and conserved regions, the comparison of these sequences can allow for differentiation at least to the genus level.32, 33 A secondary approach is ribotyping, which involves the comparative analysis of discrete-sized genome fragments generated by restriction digestion of rRNA operons.34, 35
Another method of prokaryotic identification is through matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), which is a proteotypic method that identifies proteins using peptide mass matching.36 Here, the mass-spectral pattern of intact prokaryotic cells is obtained through the analysis of ionized components, creating a unique spectrum that represents the protein profile of each sample. These spectral patterns are then compared to a database of stored profiles from known organisms, allowing for the quick and accurate identification of a species.

Bacterial Applications
For centuries, bacterial strains have been used for the production of consumable products via fermentation. In recent years, bacteria have also been commercialized for use in quality control and the production of probiotics, antibiotics, vitamins, organic acids, and biofuel. Below, we further describe various bacterial applications.
Quality Control
In the production of consumable products, it is essential that effective microbiological testing is performed to ensure the absence of contamination. The safety, reputation, and business performance of all processing companies depends on effective quality control. Strains used for quality control must have confirmed identity, viability, and purity that are backed by meticulous laboratory procedures that minimize sub-culturing. Since 1925, ATCC has been a leading provider of QC bacterial strains and has set the standard for authenticating and distributing biological materials for research and product testing. All bacterial strains authenticated and provided by ATCC have gone through both rigorous phenotypic and genotypic characterization; these procedures are described in detail in the previous chapter, “Bacterial Authentication.”
Consumable Products
Bacterial strains are commonly used in the production of various fermented food products including vinegar, pickled goods, or an assortment of dairy products. These consumable products are generated by the fermentation of compounds such as lactose or ethanol into lactic- or acetic-acid, respectively. For example, in the production of cheese and yogurt, Lactobacillus, Streptococcus, and Bifidobacterium species are commonly used for lactose fermentation. The subsequent formation of lactic acid results in a change in both the texture and flavor of the milk product. In contrast, vinegar generation requires the fermentation of ethanol by Acetobacter species, resulting in an aromatic, tangy product. Overall, as compared to many non-fermented foodstuffs, these aforementioned products are often less susceptible to spoilage and may provide additional health benefits such as improved digestion.
Probiotics
Probiotic bacterial species are thought to be beneficial to the host organism. When administered in adequate amounts, these live strains can promote healthy digestion, absorption, and a reduction in the number of enteric pathogens.37 Lactobacillus and Bifidobacterium species are the most commonly used probiotic bacterial strains and are often provided within fermented foods such as yogurt. Species within these genera have been found to naturally synthesize antibiotics, vitamins, and amino acids. Additionally, these microbes promote favorable pH levels and the absorption of nutrients including calcium, magnesium, and iron. Overall, the antimicrobial properties and health benefits associated with probiotic strains can promote proper bowel function and assist in the treatment of various conditions including gastroenteritis, colitis, and irritable bowel syndrome.
Vitamins
Many bacterial strains produce several B- and K-complex vitamins to aid in a variety of metabolic processes including DNA synthesis and the catabolism of fats, carbohydrates, or proteins. Human intestinal bacteria, for example, are known to synthesize vitamin K1, biotin (B7), folic acid (B9), niacinamide (B3), cobalamin (B12), riboflavin (B2), pantothenic acid (B5), and thiamine (B1). Many of these bacterial-produced vitamins are used in humans as coenzymes or cofactors and are considerably essential for proper metabolism. In particular, vitamin K is necessary for the formation of several blood-clotting factors in the liver. In contrast, both pantothenic acid and biotin function as coenzymes in carbohydrate oxidation and carboxylation reactions, respectively.
Antibiotics
Antibiotics are compounds used to either destroy or impede the growth of bacterial or fungal microorganisms via a variety of mechanisms such as the inhibition of protein or DNA synthesis. Many currently known antibiotics are produced by members of the bacterial class Actinomycetes; in particular, by members of the genus Streptomyces. Streptomyces species are filamentous soil bacteria that produce antibiotics through secondary metabolite pathways using various precursors such as amino acids, small fatty acids, sugars, and nucleic acids.38 Bacterial strains are believed to produce antibiotics as a means to diminish or eradicate the growth of other invading species that may be either harmful or in competition for a similar food source. Within human society, antibiotic production is critical for the treatment of infectious diseases and is often used agriculturally in the biological control of plant pathogenic microorganisms.39
Organic Acids
Various organic acids, such as citric, lactic, succinic, and gluconic acid, can be produced by bacterial species as a metabolic byproduct. Citric acid, for example, is a byproduct of the Citric Acid Cycle and is therefore important in metabolism. In contrast, the production of lactic acid often occurs as an end product to lactose fermentation. Organic acids are applicable for a variety of purposes including food preservation, emulsification, and flavoring. They have also been used in a more industrial function within buffer solutions, detergents, cosmetics, and rust removers.
Biofuels
Currently most of our energy is supplied through petroleum, coal, and natural gas deposits. These sources of energy, however, are both finite and non-renewable. Recently, efforts have been made to generate sustainable energy via the production of biofuels from commercially available products such as corn and sugar cane. However, these novel sources of energy require food to be used as fuel rather than for human sustenance. To resolve these aforementioned issues, current biofuel research is focusing on the use of bacteria in the catabolism of cellulose into desired hydrocarbon fuel compounds. The goal is to engineer bacterial strains that have metabolic pathways that can generate biofuels through the degradation of biowastes or weeds, thus creating a sustainable form of energy without diminishing the food supply.
Reporter-Labeled Strains
Fluorescence- and luminescence-based reporter-labeled strains have a diverse array of applications in the basic and applied sciences, including microbial quantification and detection, the analysis of host-pathogen interactions, drug discovery, and food testing. For example, these strains provide a unique tool for analyzing the invasion, colonization, localization, and pathogen load of a specific bacterial pathogen within host cells and tissues under different physiological conditions or stages in an infection cycle. Further, this ability to easily visualize and quantify bacteria allows for an efficient means of screening a large number of antimicrobial therapeutics and determining the growth inhibitory dose. Overall, these studies could significantly contribute to the knowledge of microbial pathogenesis, with significant implications in the development and evaluation of novel therapeutics and quality control assays.
Get a printer-friendly copy of the Bacteriology Culture Guide to keep your research on track
Download Now
Glossary
Aerobe. Organisms that require oxygen for survival. Bacterial aerobes use oxygen as the terminal electron acceptor during respiration.
Aerotolerant anaerobes. Organisms that cannot use oxygen, nor are affected by its presence.
Anaerobes. Organisms that do not require oxygen for survival. These organisms use inorganic compounds as the terminal electron acceptor during respiration.
Complex medium. An undefined medium containing unknown quantities of reagents. These media contain complex ingredients, such as yeast extract and peptone, which contain a combination of compounds in unknown proportions.
Cryopreservation. Ultra-low temperature storage of cells, tissues, embryos, or seeds. This storage is usually carried out using temperatures below -100°C.
Defibrinated blood. Whole blood from which fibrin has been separated during the clotting process.
Defined medium. Media with known proportions of all reagents. These media consist of a defined carbon and nitrogen source.
Desiccant. A hygroscopic substance that induces or sustains a state of dryness in its local vicinity in a sealed container.
Desorption. The phenomenon where a substance is released from or through a surface.
Enrichment medium. These media contain the nutrients required to support the growth of a wide array of microorganisms.
Facultative anaerobe. An organism that can survive in the presence and absence of oxygen. If given the choice, these organisms prefer the use of oxygen as it has the highest reduction potential of all terminal electron acceptors.
Fastidious. Having to do with microorganisms that have complex nutritional needs and require growth on enriched media.
Hypotonic. An environment where the water concentration is greater and solute concentration is lower on the outside of a cell. In this environment, water flows into the cell.
Isotonic. An environment where both the water and solute concentrations are the same on the inside and outside of a cell. In this environment, water flows in and out of a cell at an equal rate.
Lyophilization. A process which extracts water molecules from microbial cultures so that products remain stable and easy to store. This storage is usually carried out under low-oxygen, low-moisture conditions at 4°C.
Mesophile. An organism that grows best at moderate temperatures, between 25°C to 40°C.
Microaerophile. An organism that requires oxygen for respiration, however in lower levels than found in the atmosphere.
Obligate anaerobe. An organism that cannot survive and are often killed by the presence of oxygen.
Osmolarity. The concentration of a solution in terms of osmoles. Osmosis is the diffusion of water across a membrane from an area of higher water concentration (lower solute concentration) to lower water concentration (higher solute concentration).
Psychrophile. An extremophilic organism that is capable of growth in cold temperatures ranging from 0°C to 20°C.
Ribotyping. A method of identification involving the fingerprinting of genomic DNA restriction fragments.
Selective medium. Media used for the growth of specific microorganisms. These media are often supplemented with reagents, such as antibiotics, that prevent the growth of other microorganisms.
Thermophile. An extremophilic organism that thrives a high temperatures ranging from 45°C to 122°C.
References
- Garrity G.M. ed., et al. Bergey’s Manual of Systematic Bacteriology 2nd Edition. Springer, New York, 2001-2012.
- Adams M.H. Bacteriophages. Interscience Publishers, Inc., New York, 1959.
- Todar K. Todar’s Online Textbook of Bacteriology. Copyright; Madison, WI, 2008-2012.
- Reddy CA. Methods for general and molecular microbiology, 3rd Edition. ASM Press, Washington, D.C., 2007.
- Bauer A.W., et al. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4): 493-496, 1966.
- Middlebrook G., Cohn M.L. Bacteriology of tuberculosis: laboratory methods. Am J Public Health Nations Health. 48(7): 844-853, 1958.
- Khan J.A., et al. Nicotinamide adenine dinucleotide metabolism as an attractive target for drug discovery. Expert Opin Ther Targets. 11(5): 695-705, 2007.
- Wilkinson A., Day J., Bowater R. Bacterial DNA ligases. Mol Microbiol. 40(6): 1241-1248, 2001.
- Moir J.W. Nitrogen cycling in bacteria: Molecular analysis. Caister Academic Press, York, UK, 2011.
- The United States Pharmacopeial Convention. The United States Pharmacopeia 31-The national formulary 26. The United States Pharmacopeial Convention, Inc., Rockville, MD, 2008.
- United States Food and Drug Administration. Bacteriological Analytical Manual Edition 8. Gaithersburg, MD, 1998.
- BD Diagnositcs. Difco & BBL Manual, Manual of Microbiological Culture Media 2nd Edition. Copyright by Becton Dickinson and Company, Sparks, Maryland, 2009.
- Entner N., Doudoroff M. Glucose and gluconic acid oxidation of Pseudomonas saccharophila. J Bio Chem 196(2): 853-862, 1952.
- Giandomenico A.R., et al. The importance of sodium pyruvate in assessing damage produced by hydrogen peroxide. Free Radic Biol Med 23(3): 426-434, 1997.
- Versalovic J., ed. Manual of Clinical Microbiology 10th Edition. American Society for Microbiology, Washington D.C., 2011.
- Mazur P. The role of intracellular freezing in the death of cells cooled at supraoptimal rates. Cryobiology 14: 251-272, 1977.
- Mazur P. Cryobiology: The freezing of biological systems. Science 168: 939-948, 1970.
- Fahy G.M. The relevance of cryoprotectant “toxicity” to cryobiology. Cryobiology 23: 1-13, 1986.
- Meryman H.T. Cryoprotective agents. Cryobiology 8: 173-183, 1971.
- Rowe T.W.G., Snowman J.W. Edwards Freeze-Drying Handbook. Grand Island, New York: Edwards high Vacuum Inc., 1976.
- Heckly R.J. Principles of preserving bacteria by freeze-drying. Dev Ind Microbiol 26: 379-395, 1985.
- Ray B. Methods to detect stressed microorganisms. J Food Prot. 42: 346-355, 1979.
- Miyamoto-Shinohara Y., et al. Survival rate of microbes after freeze-drying and long-term storage. Cryobiology 41(3): 251-255, 2000.
- Clark W.A., Klein A.. The stability of bacteriophages in long-term storage at liquid nitrogen temperatures. Cryobiology 3: 68-75, 1966.
- Clark W.A., Horneland W., Klein A.G. Attempts to freeze some bacteriophages to ultralow temperatures. Appl Microbiol. 10: 463-465, 1962.
- Norman M.C. Preservation of Mycoplasmatales and L-phase variants in the American Type Culture Collection by freezing and freeze-drying. Cryobiology 10: 400-402, 1973.
- Mills C.K., Gherna R.L. Cryopreservation studies of Campylobacter. Cryobiology 25: 148-152, 1988.
- Alexander A.D., et al. Preservation of leptospiras by liquid nitrogen refrigeration. Int J Syst Bacteriol 22: 165-169, 1972.
- United States Department of Health and Human Services, Centers for Disease Control, and National Institutes of Health. Biosafety in Microbiological and Biomedical Laboratories 5th Edition. U.S. Government Printing Office, HHS Publication No. (CDC) 21-1112, Washington D.C. 2009.
- Janda J.M., Abbot S.L. Bacterial identification for publication: when is enough enough? J Clin Microbiol 40(6): 1887-1891, 2002.
- Shetty N., Hill G., Ridgway G.L. The Vitek analyser for routine bacterial identification and susceptibility testing: protocols, problems, and pitfalls. J Clin Pathol 51(4): 316-323, 1998.
- Clarridge III J. Impact of 16S rRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clin Microbiol Rev 17(4): 840-862, 2004.
- Janda J.M., Abbott S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45(9): 2761-2761, 2007.
- Bruce J. Automated system rapidly identifies and characterizes micro-organisms in food. Food Technology 50: 77-78, 1996.
- Woese C. Bacterial evolution. Microbiology Review 51: 221-271, 1987.
- Demirev P.A., et al. Microorganism identification by mass spectrometry and protein database searches. Anal Chem 71(14): 2732-2738, 1999.
- Nagpal R., et al. Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiol Lett Epub ahead of print, 2012.
- Chater K.F. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos Trans R Soc Lond B Biol Sci. 361: 761-768, 2006.
- Raaijmakers J.M., Vlami M., de Souza J.T. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81(1- 4): 537-547, 2002.
